Latent functional diversity may accelerate microbial community responses to temperature fluctuations
Figures
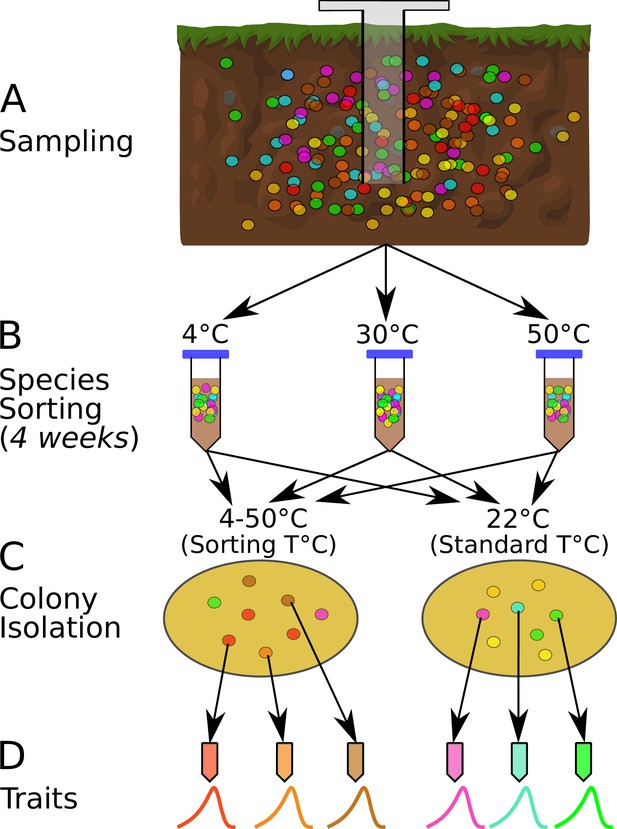
The species sorting experiment.
(A) Different bacterial taxa (colored circles) sampled from the soil community. (B) Samples maintained at 4, 10, 21, 30, 40, and 50°C (only three temperatures shown for illustration), allowing species sorting for 4 weeks. (C) Soil washes from each core plated out onto agar and grown at both the sorting temperature and 22°C (standard temperature) to allow further species sorting and facilitate isolation (next step). (D) The six most abundant (morphologically different) colonies from each plate were picked, streaked, and isolated, and their physiological and life history traits measured. The curves represent each strain’s unique unimodal response of growth rate to temperature.
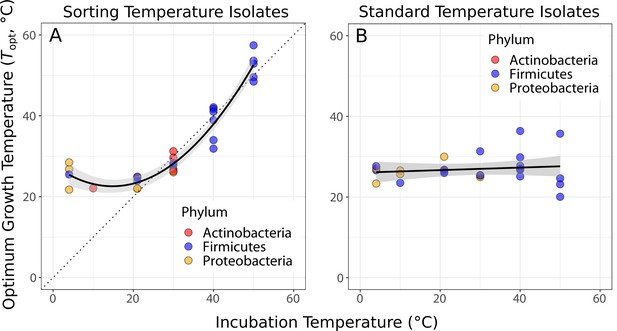
Species sorting of soil bacteria driven by temperature change.
(A) Thermal optima of growth rate closely match sorting temperature for the isolates from those temperatures (black line: quadratic linear regression, p<0.0001, R2 = 0.94, = 28). Note that the prediction bounds at three lowest temperatures do not include the 1:1 (dashed) line. (B) No significant association between incubation temperature and thermal optima for standard temperature isolates (simple linear regression, p = 0.488, R2 = 0.02, = 26). These results show that species sorting can act upon latent diversity to select for isolates adapted to different temperature conditions (A), but that isolates maladapted to the sorting conditions can re-emerge (be resuscitated) under the appropriate conditions (B).
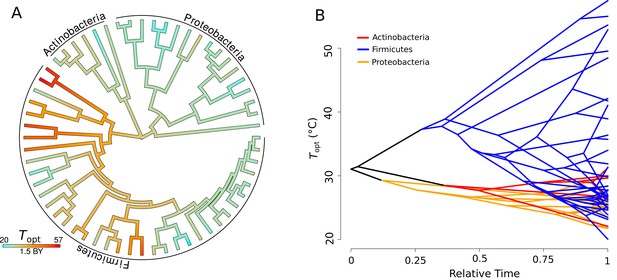
Evolution of .
(A) Ancestral trait reconstruction of visualized on a tree, from lower temperatures in cyan, to higher temperatures in red, with time given in billions of years (BY). All of the higher temperature (40–50°C) isolates belong to the phylum Firmicutes. (B) Projection of the phylogenetic tree into the trait space (y-axis), over relative time (x-axis) since divergence from the root. The clades representative of each phylum are colored on the projection (Actinobacteria, red; Firmicutes, blue; Proteobacteria, yellow).
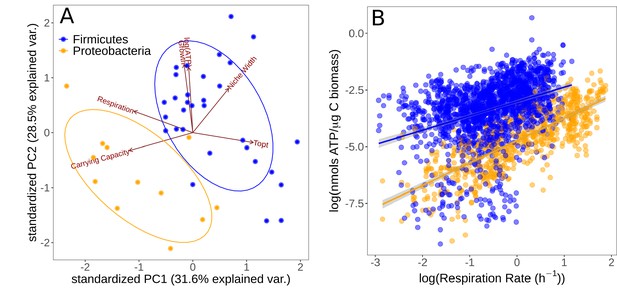
Partitioning of growth strategies between phyla.
(A) Principal components analysis (PCA) on life history traits, colored by phylum. Relative to each other, Firmicutes (blue) tend to be specialists, Proteobacteria (orange) tend to be specialists. (B) ATP content of cultures is associated with the respiration rate. Firmicutes show a sublinear scaling relationship of ATP with respiration rate (scaling exponent = 0.60 ± 0.07), while Proteobacteria display an approximately linear scaling relationship (scaling exponent = 0.99 ± 0.06). The same color scheme is shared by both sub-plots.
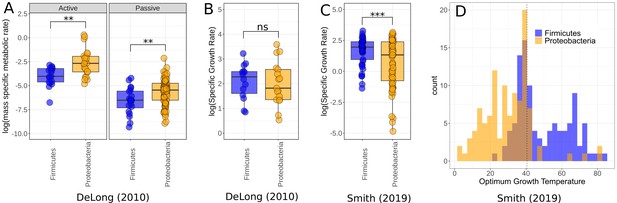
Comparison of Firmicutes and Proteobacteria in meta-analysis datasets.
(A) Dataset used by DeLong et al., 2010 shows significantly higher active (n = 39) and passive (n = 108) metabolic rates for Proteobacteria than Firmicutes. Significance determined by Wilcoxon rank-sum tests – ns, p≥0.05; *p<0.05; **p<0.01; ***p<0.001. (B) The growth rate data used by DeLong et al., 2010 shows no significant difference between the phyla (n = 31). (C) The growth rate data from Smith et al., 2019 does show significantly increased growth rates for Firmicutes over Proteobacteria however (n = 135). (D) Distribution of Firmicutes and Proteobacteria from Smith et al., 2019. Proteobacteria account for a large proportion of the low-temperature strains, while Firmicutes dominate the high temperatures. Dotted line marks 40.5°C, a cut-off between mesophiles and thermophiles (Smith et al., 2019).
Tables
Details of time tree calibration nodes.
We constrained the time calibration of our RAxML tree based on estimated divergence times from TimeTree (Kumar et al., 2017).
| Taxa A | Taxa B | Min divergence time (MYA) | Max divergence time (MYA) |
|---|---|---|---|
| Bacteria | Archaea | 4290 | - |
| Pseudomonas | Bacillus | 3100 | 3254 |
| Pseudomonas | Labrys | 1053 | 3135 |
| Pseudomonas | Collimonas | 1053 | 3135 |
| Collimonas | Variovorax | 1271 | - |
| Arthrobacter | Streptomyces | 1420 | 1870 |
| Arthrobacter | Bacillus | 3100 | 3254 |
| Bacillus | Brevibacillus | 1734 | 2398 |
List of revivable strains and GenBank accession numbers.
Strain codes follow XX_YY_ZZ naming convention, where XX is the incubation temperature, YY is the isolation temperature, and ZZ is a numeric designator for the specific isolate. RT = room temperature (22°C, termed ‘standard temperature’). All 16S sequences are archived on NCBI’s GenBank with the accession numbers indicated.
| Strain | Accession no. | Phylum | Class | Order | Family | Genus |
|---|---|---|---|---|---|---|
| 04_04_02 | ON804144 | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas |
| 04_04_04 | ON804145 | Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteraceae | Collimonas |
| 04_04_05 | ON804146 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 04_04_06 | ON804147 | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas |
| 04_RT_01 | ON804148 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 04_RT_02 | ON804149 | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas |
| 04_RT_03 | ON804150 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 04_RT_05 | ON804151 | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas |
| 10_10_06 | ON804152 | Actinobacteria | Actinobacteria | Micrococcales | Micrococcaceae | Arthrobacter |
| 10_RT_01 | ON804153 | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas |
| 10_RT_02 | ON804154 | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas |
| 10_RT_03 | ON804155 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 21_21_01 | ON804156 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 21_21_02 | ON804157 | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas |
| 21_21_04 | ON804158 | Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteraceae | Collimonas |
| 21_21_05 | ON804159 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 21_21_06 | ON804160 | Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas |
| 21_RT_01 | ON804161 | Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteraceae | Collimonas |
| 21_RT_02 | ON804162 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 21_RT_03 | ON804163 | Firmicutes | Bacilli | Bacillales | Paenibacillaceae | Paenibacillus |
| 21_RT_04 | ON804164 | Proteobacteria | Gammaproteobacteria | Xanthomonadales | Rhodanobacteraceae | Dyella |
| 21_RT_05 | ON804165 | Actinobacteria | Actinobacteria | Corynebacteriales | Nocardiaceae | Nocardia |
| 21_RT_06 | ON804166 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 30_30_01 | ON804167 | Actinobacteria | Actinobacteria | Streptomycetales | Streptomycetaceae | Streptomyces |
| 30_30_02 | ON804168 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 30_30_03 | ON804169 | Actinobacteria | Actinobacteria | Streptomycetales | Streptomycetaceae | Streptomyces |
| 30_30_04 | ON804170 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 30_30_05 | ON804171 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 30_30_06 | ON804172 | Actinobacteria | Actinobacteria | Streptomycetales | Streptomycetaceae | Streptomyces |
| 30_30_07 | ON804173 | Actinobacteria | Actinobacteria | Streptomycetales | Streptomycetaceae | Streptomyces |
| 30_30_08 | ON804174 | Proteobacteria | Alphaproteobacteria | Rhizobiales | Xanthobacteraceae | Labrys |
| 30_RT_01 | ON804175 | Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | Variovorax |
| 30_RT_02 | ON804176 | Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | Variovorax |
| 30_RT_03 | ON804177 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 30_RT_04 | ON804178 | Proteobacteria | Gammaproteobacteria | Xanthomonadales | Rhodanobacteraceae | Dyella |
| 30_RT_05 | ON804179 | Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | Variovorax |
| 30_RT_06 | ON804180 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 40_40_01 | ON804181 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 40_40_02 | ON804182 | Firmicutes | Bacilli | Bacillales | Paenibacillaceae | Cohnella |
| 40_40_03 | ON804183 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 40_40_04 | ON804184 | Firmicutes | Bacilli | Bacillales | Planococcaceae | Rummeliibacillus |
| 40_40_05 | ON804185 | Firmicutes | Bacilli | Bacillales | Paenibacillaceae | Cohnella |
| 40_40_06 | ON804186 | Firmicutes | Bacilli | Bacillales | Planococcaceae | Viridibacillus |
| 40_RT_01 | ON804187 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 40_RT_02 | ON804188 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 40_RT_03 | ON804189 | Firmicutes | Bacilli | Bacillales | Planococcaceae | Viridibacillus |
| 40_RT_04 | ON804190 | Firmicutes | Bacilli | Bacillales | Planococcaceae | Viridibacillus |
| 40_RT_05 | ON804191 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 40_RT_06 | ON804192 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 50_50_01 | ON804193 | Firmicutes | Bacilli | Bacillales | Planococcaceae | Rummeliibacillus |
| 50_50_02 | ON804194 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Anoxybacillus |
| 50_50_03 | ON804195 | Firmicutes | Bacilli | Bacillales | Paenibacillaceae | Brevibacillus |
| 50_50_04 | ON804196 | Firmicutes | Bacilli | Bacillales | Paenibacillaceae | Brevibacillus |
| 50_50_05 | ON804197 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 50_50_06 | ON804198 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Anoxybacillus |
| 50_RT_01 | ON804199 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 50_RT_02 | ON804200 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 50_RT_03 | ON804201 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 50_RT_04 | ON804202 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |
| 50_RT_06 | ON804203 | Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus |





