Drivers of species knowledge across the tree of life
Figures
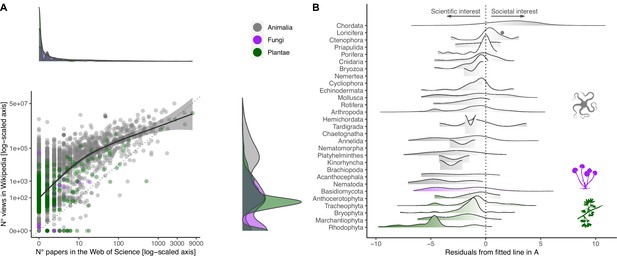
Relationship between societal and scientific interest across the eukaryotic Tree of Life.
(A) Relationship between number of views in Wikipedia (popular interest) and number of papers in the Web of Science (scientific interest) for each species. Both axes are log-scaled to ease visualization. Density functions are provided for both scientific (above scatter plot) and societal interests (right of scatter plot) to illustrate the distribution of values. Color coding refers to the three realms of Animalia, Fungi, and Plantae. The regression line is obtained by fitting a Gaussian generalized additive model through the data (F1,3017 = 2497.5; p<0.001). The farther away a dot is from the fitted line, the more the attention is unbalanced toward either scientific (negative residuals) or societal interest (positive residuals). (B) Distribution of negative and positive residuals (from the regression line in A) across the species sampled for each Phylum/Division. Phyla/Divisions with only one sampled species are represented with dots.
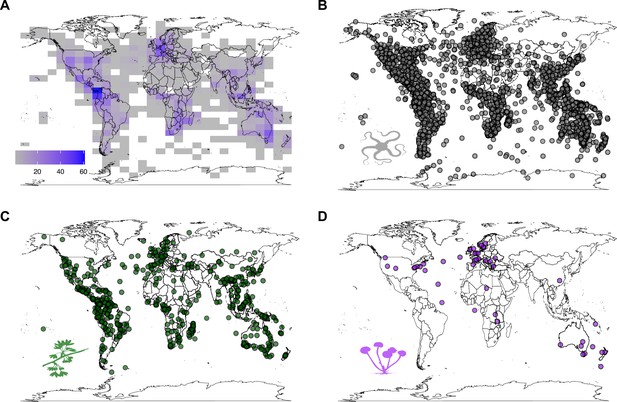
Global distribution of the sampled species.
Maps are based on the centroid of the distribution of each species, hence only species for which distribution points were available in the Global Biodiversity Information Facility are depicted (89%, n = 2687). (A) Density of distribution centroids across all groups; (B) distribution of centroids for animals; (C) distribution of centroids for plants; and (D) distribution of centroids for fungi.
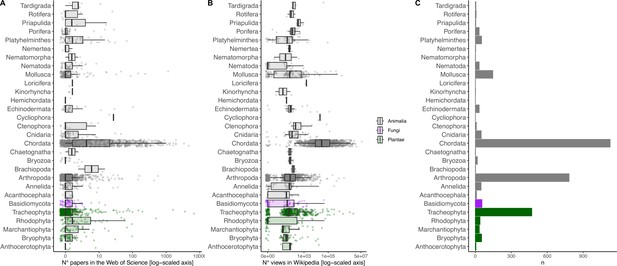
Breakdown of scientific (A) and societal (B) interest by phylum, as well as the number of sampled species (C).
Jittered points are the actual values; boxplots summarize median and quantiles. Both axes are log-scaled to ease visualization.

Influence of species-level traits (blue) and cultural factors (red) on relative scientific and societal interest for different taxa.
Forest plots summarize the estimated parameters based on Gaussian linear mixed models testing the relationship between residuals from the regression line in Figure 1A and species-level traits. Positive residuals indicate species with a greater popular than scientific interest, residuals close to zero indicate species with a balanced scientific and societal interest, and negative residuals indicate species with a greater scientific than societal interest (Figure 1B). Baseline levels for multilevel factor variables are: Domain (Multiple) and International Union for Conservation of Nature (IUCN) (Unknown). Error bars mark 95% confidence intervals. Variance explained is reported as marginal R2, that is, those explained by fixed factors. Asterisks (*) mark significant effects (α = 0.01). Estimated regression parameters are provided in Supplementary file 1a.

Influence of species-level traits (blue) and cultural factors (red) on the scientific and societal interest across the eukaryotic Tree of Life.
(A) Forest plots summarize the estimated parameters based on negative binomial generalized linear mixed models (Equation 1). Baseline levels for multilevel factor variables are: Domain (Multiple) and IUCN (Unknown). Error bars mark 95% confidence intervals. Variance explained is reported as marginal R2, that is, those explained by fixed factors. Asterisks (*) mark significant effects (α = 0.01). Exact estimated regression parameters and p-values are provided in Supplementary file 1a. (B) Outcomes of the variance partitioning analysis, whereby we partitioned out the relative contribution of species-level traits (blue) and culture factors (red). Joint explained variance (Species + Culture) is highlighted in purple. Unexplained variance is the amount of unexplained variance after considering the contribution of random factors related to species’ taxonomy and biogeographic origin (as obtained via conditional R2).
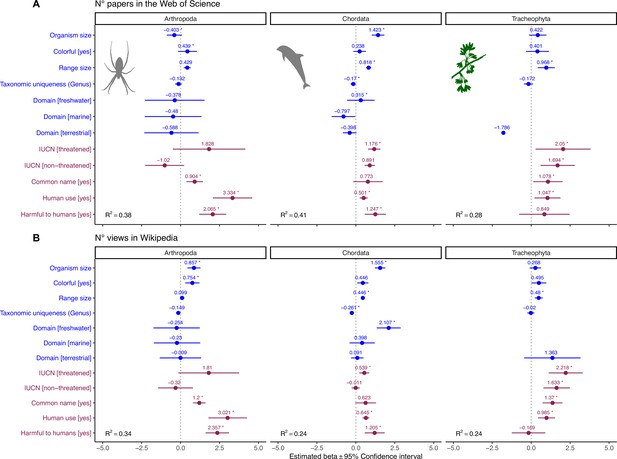
Influence of species-level traits (blue) and cultural factors (red) on scientific (A) and societal (B) interest for Arthopoda, Chordata, and Tracheophyta.
Forest plots summarize the estimated parameters based on negative binomial generalized linear mixed models (Equation 2). Baseline levels for multilevel factor variables are: Domain (Multiple) and IUCN (Unknown). Error bars mark 95% confidence intervals. Variance explained is reported as marginal R2, that is, those explained by fixed factors. Asterisks (*) mark significant effects (α = 0.01). Estimated regression parameters and p-values are provided in Supplementary file 1b (A) and Supplementary file 1c (B).
Additional files
-
Supplementary file 1
Estimated parameters for the regression models.
(a) Estimated regression parameters for the full models. (b) Estimated regression parameters for the subset models modeling the number of papers in the Web of Science. (c) Estimated regression parameters for the subset models modeling the number of views in Wikipedia.
- https://cdn.elifesciences.org/articles/88251/elife-88251-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/88251/elife-88251-mdarchecklist1-v1.docx





