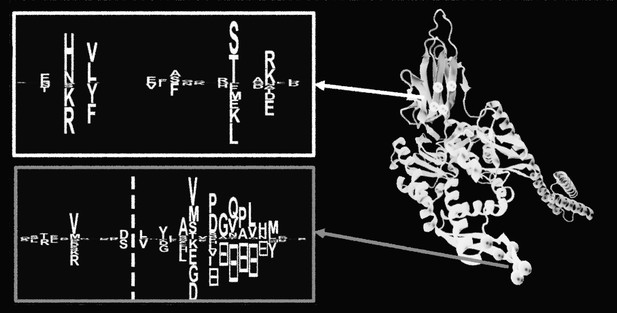
Stylized version of two inferred patterns extracted from a chaperone protein using RBM. Adapted from images provided by Tubiana et al. (CC BY 4.0)
Almost every process that keeps a cell alive depends on the activities of several proteins. All proteins are made from chains of smaller molecules called amino acids, and the specific sequence of amino acids determines the protein’s overall shape, which in turn controls what the protein can do. Yet, the relationships between a protein’s structure and its function are complex, and it remains unclear how the sequence of amino acids in a protein actually determine its features and properties.
Machine learning is a computational approach that is often applied to understand complex issues in biology. It uses computer algorithms to spot statistical patterns in large amounts of data and, after 'learning' from the data, the algorithms can then provide new insights, make predictions or even generate new data.
Tubiana et al. have now used a relatively simple form of machine learning to study the amino acid sequences of 20 different families of proteins. First, frameworks of algorithms –known as Restricted Boltzmann Machines, RBM for short – were trained to read some amino-acid sequence data that coded for similar proteins. After ‘learning’ from the data, the RBM could then infer statistical patterns that were common to the sequences. Tubiana et al. saw that many of these inferred patterns could be interpreted in a meaningful way and related to properties of the proteins. For example, some were related to known twists and loops that are commonly found in proteins; others could be linked to specific activities. This level of interpretability is somewhat at odds with the results of other common methods used in machine learning, which tend to behave more like a ‘black box’.
Using their RBM, Tubiana et al. then proposed how to design new proteins that may prove to have interesting features. In the future, similar methods could be applied across computational biology as a way to make sense of complex data in an understandable way.




