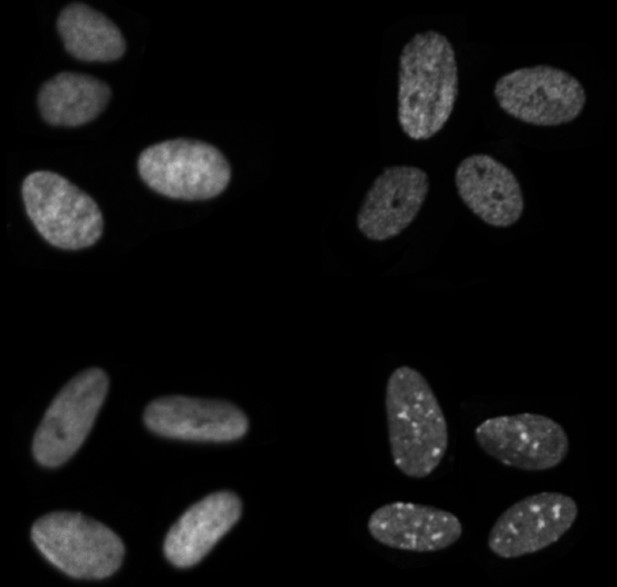
Cells stained for four different histone modifications, which can be used by MIEL to determine the epigenetic state of the cell. Image credit: Farhy et al. (CC BY 4.0)
Each cell contains a complete copy of a person’s genes coded in their DNA. However, for a cell to perform its specific role, it only needs a small fraction of this genetic information. The mechanisms that control which genes a cell is using fall under the umbrella of ‘epigenetics’ (meaning beyond genetics). These mechanisms involve changes in the way that DNA is organized inside the cell nucleus and changes in how accessible different parts of the genome are to various cellular components.
DNA is long and fragile so, to maintain its integrity, it is wrapped around protein complexes called histones. Adding chemical modifications to histones is one of the main epigenetic mechanisms that cells use to regulate which genes are turned on and off. Several methods allow researchers to read patterns of histone modification and use this information to derive what state a cell is in and how it might behave. Improving these methods is of particular interest in drug development, where this information could reveal the effects, and side-effects, of new treatments. Unfortunately, existing techniques are costly in both time and money, and they are not well suited to analyzing epigenetic changes caused by the large numbers of compounds tested during drug development.
To overcome this barrier, Farhy et al. have developed a new system called ‘Microscopic Imaging of the Epigenetic Landscape’ (MIEL for short). The system allows them to quickly analyze the epigenetic changes caused by each of a large number of different chemical compounds when they are used on cells. MIEL tags different histone modifications by staining each with a different color, and then uses automated microscopy to produce images. It then extracts information from these images using advanced image analysis tools. The changes induced by different drugs can then be compared and categorized using machine learning algorithms.
To test the MIEL system, Farhy et al. grew brain cancer cells (derived from human tumors) in the lab, and treated them with compounds that target proteins involved in histone modifications. Using their newly created pipeline, Farhy et al. were able to identify the unique epigenetic changes caused by these compounds, and train the system to correctly predict which type of drug the cells had been treated with. In a different set of experiments Farhy et al. demonstrate the utility of their new pipeline in identifying drugs that induce a set of epigenetic changes associated with a reduced ability to regrow tumors.
This new system could help screen thousands of compounds for their epigenetic effects, which could aid the design of new treatments for many diseases.




