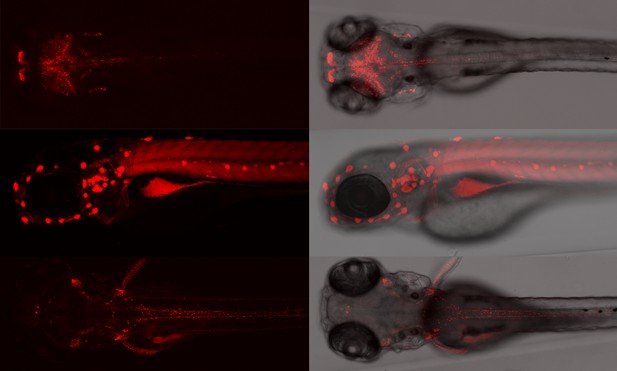
A zebrafish in which a specific protein has been deactivated and tagged (in red). Image credit: Ichino et al. (CC BY 4.0)
The human genome counts over 20,000 genes, which can be turned on and off to create the proteins required for most of life processes. Once produced, proteins need move to specific locations in the cell, where they are able to perform their jobs. Despite striking scientific advances, 90% of human genes are still under-studied; where the proteins they code for go, and what they do remains unknown.
Zebrafish share many genes with humans, but they are much easier to manipulate genetically. Here, Ichino et al. used various methods in zebrafish to create a detailed ‘catalogue’ of previously poorly understood genes, focusing on where the proteins they coded for ended up and the biological processes they were involved with.
First, a genetic tool called gene-breaking transposons (GBTs) was used to create over 1,200 strains of genetically altered fish in which a specific protein was both tagged with a luminescent marker and unable to perform its role. Further analysis of 204 of these strains revealed new insight into the role of each protein, with many having unexpected roles and localisations. For example, in one zebrafish strain, the affected gene was similar to a human gene which, when inactivated, causes severe muscle weakness. These fish swam abnormally slowly and also had muscle problems, suggesting that the GBT fish strains could ‘model’ the human disease.
This work sheds new light on the role of many previously poorly understood genes. In the future, similar collections of GBT fish strains could help researchers to study both normal human biology and disease. They could especially be useful in cases where the genes responsible for certain conditions are still difficult to identify.




