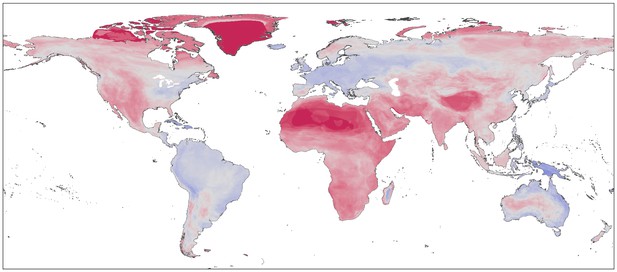
A world map with regions color-coded between blue and red. Red regions are areas where the number of species predicted to be susceptible to SARS-CoV-2 based on Mollentze et al.’s new approach (which uses species similarity) is greater than the expected number would be if species with susceptibility were distributed evenly across the globe. Blue regions are areas with fewer species predicted to be susceptible to SARS-CoV-2 than if susceptible species were evenly distributed. The map demonstrates that geographic areas containing more species predicted to be susceptible to SARS-CoV-2 (areas in red) are widespread. Local information beyond susceptibility – such as rate of contact with humans – will need to be considered to narrow down the number of species at risk to the point where monitoring of specific species becomes practical.. Image credit: Mollentze et al. (CC BY-SA 4.0)
The COVID-19 pandemic affects humans, but also many of the animals we interact with. So far, humans have transmitted the SARS-CoV-2 virus to pet dogs and cats, a wide range of zoo animals, and even wildlife. Transmission of SARS-CoV-2 from humans to animals can lead to outbreaks amongst certain species, which can endanger animal populations and create new sources of human infections. Thus, careful monitoring of animal infections may help protect both animals and humans.
Identifying which animals are susceptible to SARS-CoV-2 would help scientists monitor these species for outbreaks and viral circulation. Unfortunately, testing whether SARS-CoV-2 can infect different species in the laboratory is both time-consuming and expensive. To overcome this obstacle, researchers have used computational methods and existing data about the structure and genetic sequences of ACE2 receptors – the proteins on the cell surface that SARS-CoV-2 uses to enter the cell – to predict SARS-COV-2 susceptibility in different species. However, it remained unclear how accurate this approach was at predicting susceptibility in different animals, or whether their correct predictions indicated causal links between ACE2 variability and SARS-CoV-2 susceptibility.
To assess the usefulness of this approach, Mollentze et al. started by using data on the ACE2 receptors from 96 different species and building a machine learning model to predict how susceptible those species might be to SARS-CoV-2. The susceptibility of these species had either been observed in natural infections – in zoos, for example – or had been assessed in the laboratory, so Mollentze et al. were able to use this information to determine how good both their model and previous approaches based on the sequence of ACE2 receptors were.
The results showed that while the model was quite accurate (it correctly predicted susceptibility to SARS-CoV-2 about 80% of the time), its predictions were based on regions of the ACE2 receptors that were not known to interact with the virus. Instead, the regions that the machine learning model relied on were ones that tend to vary more the more distantly related two species are. This indicates that existing computational approaches are likely not relying on information about how ACE2 receptors interact with SARS-CoV-2 to predict susceptibility. Instead, they are simply using information on how closely related the different animal species are, which is much easier to source than data about ACE2 receptors.
Indeed, the sequences of the ACE2 receptors in many species are unknown and the species for which this information is available come only from a few geographic areas. Mollentze et al. also showed that limiting the predictions about susceptibility to these species could mislead scientists when deciding which species and geographic areas to surveil for possible viral circulation.
Instead, it may be more effective and cost-efficient to use animal relatedness to predict susceptibility to SARS-CoV-2. This makes it possible to make predictions for nearly all mammals, while being just as accurate as models based on ACE2 receptor data. However, Mollentze et al. point out that this approach would still fail to narrow down the number of animals that need to be monitored enough for it to be practical. Considering additional factors like how often the animals interact with humans or how prone they are to transmit the virus among themselves may help narrow it down more. Further research is therefore needed to identify the best multifactor approaches to identifying which animal populations should be monitored.




