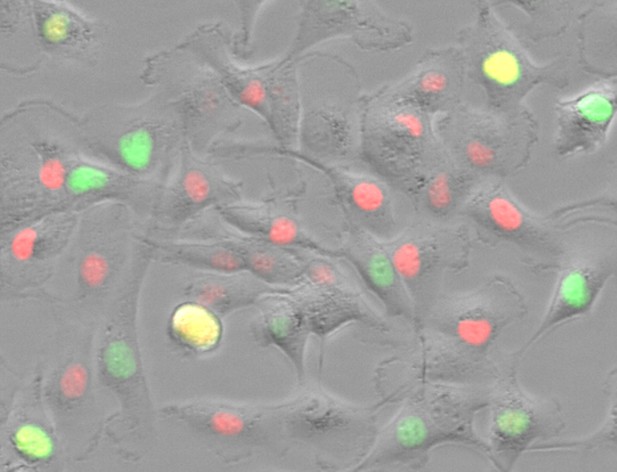
Blood cancer cells grown on a nano-material-coated surface expressing a fluorescent marker to highlight different cell cycle phases. Image credit: Hayatigolkhatmi et al. (CC BY 4.0)
Cells regularly grow and divide through a process called the cell cycle. It includes rest periods where no growth or division occurs. When the cells are ready to divide, they duplicate their DNA, so each new cell gets a complete set of instructions. Finally, the cell splits into two new cells through a process called cytokinesis. This whole process can take hours or days to complete, depending on the cell type. Many things can go wrong during these processes, impairing healing or causing tumor formation. Learning more about these processes could help scientists better understand healing and diseases like cancer.
Emerging imaging and data analysis tools allow scientists to observe cell-growth processes and watch errors as they occur. But, doing so requires sophisticated equipment and can be time and labor-intensive. Especially, if scientists are trying to track the cell cycle in a large number of cells. It can also be challenging to track free-moving cells, like blood or immune cells. New tools and techniques are needed to help scientists overcome these challenges.
Hayatigolkhatmi, Soriani, Soda et al. developed a method in which a sticky surface is used to grow blood cancer cells that allows them to observe the cell cycle in large numbers of the cells at the same time. In the experiments, blood cancer cells were grown on a nano-material-coated surface that kept the usually free-floating cells still. The team compared gene expression in the cells before and after raising them on the surface to confirm that confining the cells did not alter their gene expression or disrupt their normal life cycle. Then, the researchers developed machine learning software that monitors the cell cycle in hundreds of individual cells, quantifies cell cycle phases and analyzes data with minimal human intervention. Usually, it would take a scientist 40-50 hours to oversee the cell cycle in a single experimental condition. This time was reduced to approximately 2 hours for a complete experiment using their pipeline. Finally, they validated their tools by monitoring different types of cancer cells under various treatment conditions.
The tools developed by Hayatigolkhatmi, Soriani, Soda et al. provide researchers with a fast, easy and cost-effective tool for studying the cell cycle. It could help scientists study early development and how cells differentiate, grow or age. It could also be helpful for scientists studying cancer and how to treat it or scientists studying the healing process.




