By Heather Staines, Hypothesis; Nathan Lisgo and Naomi Penfold, eLife
The teams at eLife and Hypothesis are pleased to announce that annotation capability is now live on the new eLife platform. The development completed by our collaboration lets publishers seamlessly integrate moderated annotation layers into their content, which lets users view and, after logging in with their ORCID credentials, create private and public annotations, page notes and highlights. In addition, publishers are now able to customise aspects of the annotation user interface (UI) to more gracefully blend its functionality into the look and feel of their sites.
To get started with annotations in eLife, users should register with their ORCID id and start annotating by clicking the ‘annotations’ link at the top of any article or highlighting a section of text.
How annotation works with Hypothesis
Hypothesis is an open-source annotation technology, based on the World Wide Web consortium (W3C) standards, that enables rich, interconnected conversations across all the content on the web. The publication of annotations as a web standard by the W3C in February 2017 paved the way to bringing digital annotation natively to browsers, fulfilling an original vision for networked information laid out by Vannevar Bush in 1945. By committing to a web standard, the progress we make today should survive independently of our organisations. As implemented on eLife, the tool enables users to highlight their own selections of text and create public or private notes that apply to the whole article as well as annotations on selected text. Users can find all their highlights and annotations in one place on their profile page and navigate from each note back to where they made it.
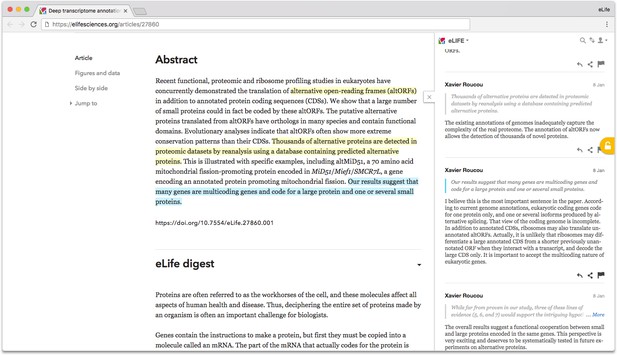
An author adds comments inline to an eLife article. Source: https://doi.org/10.7554/eLife.27860.
Hypothesis uses a standard client-server architecture, where code embedded in a web page adds client annotation tools into the user experience when a web page is loaded in the browser. As a user highlights text and creates annotations, the embedded annotation client records this such that the next user visiting an annotated page can read any public annotations, while authenticated users can also reply to existing annotations or create their own private annotations. This is all thanks to Hypothesis’ application programming interface (API)-driven infrastructure.
Annotations are attached to pages based on a user’s text highlights, and maintain that association even as a page changes over time using what Hypothesis calls "fuzzy anchoring". Fuzzy anchoring involves three different ways of recording an anchor's relationship to a document's text, which are evaluated through a four-step process to ensure the greatest possibility of accurate attachment despite changes to the text within or outside the anchor. Read more about fuzzy anchoring and its development from other anchoring techniques.
Beyond anchoring, Hypothesis annotations are also available across different versions of the same document, based on a process of mapping document equivalence with URLs, DOIs, PDF fingerprints and other document metadata. For example, an annotation made on a PDF version of a document — even on a local computer — can show up on an HTML version of the same document. Read more about document equivalence and how annotations are mapped to different document formats.
Bringing annotation to the scientific literature
More publishers are starting to view discussions attached to their publications via commenting and annotation systems as a complement to the article’s Version of Record. They want to feel confident that they can apply their own editorial standards to these annotations, just as they do with their articles themselves. With this in mind, the eLife and Hypothesis teams worked to define and implement a set of core features that would give publishers an increased level of control over their annotations and the ability to:
- streamline the integration of the annotation tool and the publisher’s own user account and authentication systems
- moderate their users’ annotations to ensure civil and on-topic dialogue
- customise the annotation tool to seamlessly blend into the flow of the publisher’s user experience
This was a highly rewarding project to work on... throughout we knew we were contributing to something greater than an additional feature on our site.
A key component of the development collaboration was to evolve Hypothesis’ authentication capabilities so they could integrate with eLife's ORCID-based system. This makes it possible for eLife users to annotate using their eLife user account, instead of requiring a separate log in to Hypothesis. In parallel, Hypothesis has made a core change to move from cookie-based authentication to a more flexible and secure system based on the widely-used OAuth standard. Overall, these developments enable Hypothesis to integrate with a much wider variety of authentication and identity solutions.
To support eLife's requirement to steward the quality of discussion in annotations within its community, the collaboration also developed a moderation process for annotations. Users can now flag improper or suspect annotations for action by administrators.
The third major outcome of eLife’s and Hypothesis' collaboration was the ability for publishers to have greater control over the function and appearance of the annotation client embedded in their site or platform. The Hypothesis client can now be configured to use a ‘clean’, customised user-interface theme to integrate more seamlessly with publisher sites, and to act in different ways to support publisher needs and preferences. It’s as simple as setting configuration values in the javascript used to load the client. The eLife user interface for Hypothesis was designed with extensive user testing to give the best experience possible. It was developed through close collaboration between eLife’s and Hypothesis’ user experience design teams with the aim of streamlining the annotation experience, removing interface distractions and better supporting first-time users.
Evolving the notion of comments at eLife
Implementing Hypothesis on eLife allows our authors and readers to interact in a new way, including seeing new metrics on which part of an article is most interesting to readers. Prior to this, readers could post public comments below eLife articles using Disqus, a widely used third-party commenting tool. It was important to us that this valuable scientific discussion was preserved as we moved technologies, and so we developed a mechanism to migrate and convert the comments from Disqus into Hypothesis page notes on eLife. In doing so, we took care to conserve the complex content of these comments as they were converted from HTML to markdown, the language that the Hypothesis annotation engine renders. This was not always simple, especially for mathematical equations.
We also wanted to transfer the key metadata — comment author and posting date — as faithfully as possible. The basic migration process is possible using the Hypothesis API, so any content publishers wishing to migrate should be able to do so without needing to work directly with Hypothesis. For us, we chose to amend the page note posting date to reflect the original comment posting date, which required working with the Hypothesis team to update its database. Now authors of previous Disqus comments can reclaim these comments when they first log in to their eLife user account, as long as they are using the email address used when submitting the original comment. Other publishers may choose to programmatically include the metadata within the content of the page note instead, thus avoiding this additional work.
“This was a highly rewarding project to work on: so many different voices were involved in bringing this to fruition, our contributions went directly into a codebase that will be used by others, and throughout we knew we were contributing to something greater than an additional feature on our site.” — Nathan Lisgo, Senior Drupal Developer, eLife
“Working with the eLife team on this project was an incredibly rewarding and collaborative experience. We couldn’t have asked for a better team to partner with on our first publisher integration of Hypothesis. The moderation, customisation, authentication and groups work our teams completed are open and available for any Hypothesis user, and we will continue to build Hypothesis to forward our mission of enabling conversations over the world’s knowledge.” — Arti Walker-Peddakotla, Senior Product Manager, Hypothesis
Open for all
These technical innovations were possible thanks to close collaboration between the eLife and Hypothesis teams, who worked together over most of 2017 as eLife was also developing major changes to its platform. This model of collaborative, open-source, standards-based development addressed the very practical needs surfaced by the eLife team, while also ensuring its outcomes could be generalised for use by everyone. All developments were made to the master branch of the Hypothesis source code and are open for the wider community to adopt and extend. eLife and Hypothesis will continue to work to shape best practices for annotation on eLife and the continuing evolution of Hypothesis technologies.
To use the Hypothesis API, see http://h.readthedocs.io/en/latest/api/.
Publishers interested in integrating the Hypothesis annotation tool into their platform should direct enquiries to Heather Staines (heather [at] hypothes [dot] is).
Do you have an idea or innovation to share? Apply now for the eLife Innovation Sprint (May 2018) or send a short outline for a Labs blogpost to innovation [at] elifesciences [dot] org.
For the latest in innovation, eLife Labs and new open-source tools, sign up for our technology and innovation newsletter. You can also follow @eLifeInnovation on Twitter.




