Molecular architecture of human polycomb repressive complex 2
Figures
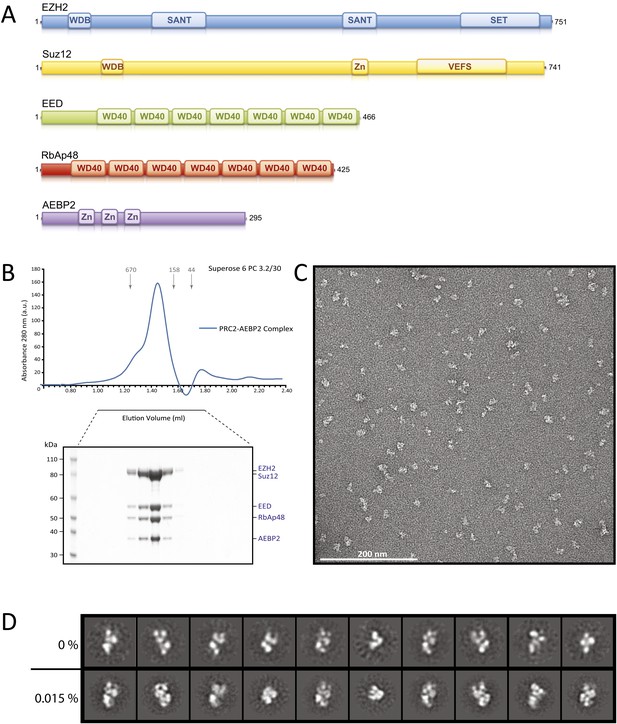
Reconstitution of the human PRC2-AEBP2 Complex.
(A) Schematic representation of the components of the human PRC2 Complex and AEBP2. (B) Size-exclusion chromatography of recombinant PRC2-AEBP2 complex and corresponding SDS-PAGE separation stained with Coomassie brilliant blue. The molecular mass of the recombinant complex is ∼275 kDa. Arrows and numbers indicate elution markers in the size-exclusion chromatography experiments and their molecular masses (in kilodaltons), respectively (a.u.: arbitrary unit). (C) Negative-stain EM of the recombinant PRC2-AEBP2 complex. Individual particles have an elongated shape with a length of ∼16 nm and a thickness of ∼7 nm. Bar: 200 nm. (D) Comparison between representative reference-free 2D class averages from non cross-linked (0%) and mildly cross-linked (0.015% glutaraldehyde) particles of the PRC2-AEBP2 complex.
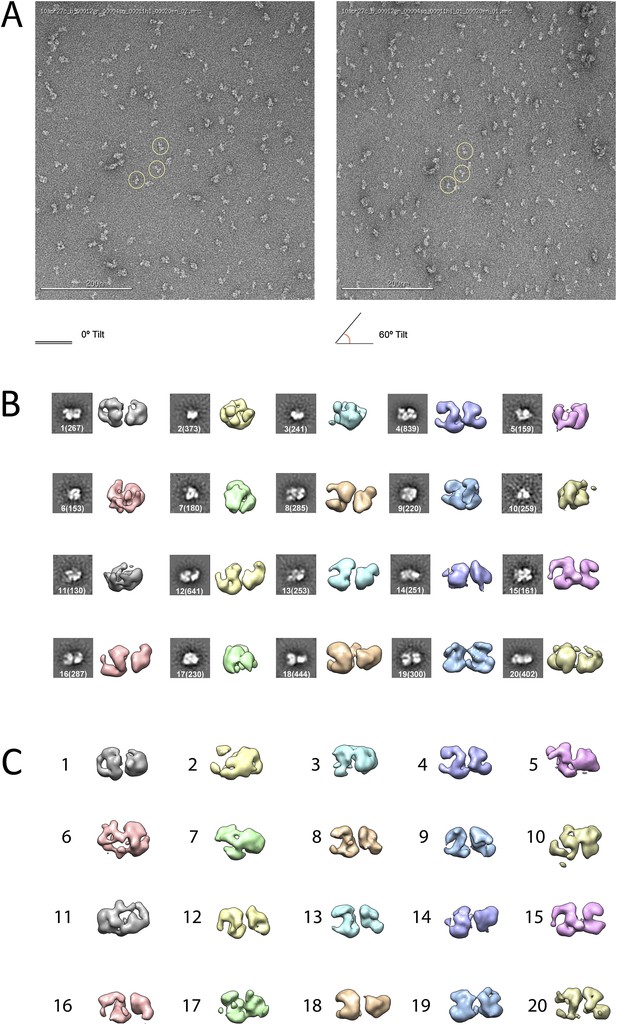
Ab initio random conical tilt reconstruction of the human PRC2-AEBP2 complex.
(A) Representative untilted and 60° tilt-pair micrographs (80,000× magnification). Corresponding particles pairs indicated by yellow circles. (B) RCT Volumes aligned to each of the 20 corresponding reference free class averages (from 6075 particles in the 0° micrographs, each class containing between 150 and 800 particles, as indicated in parentheses). (C) Alignment of the RCT volumes with respect to each other.
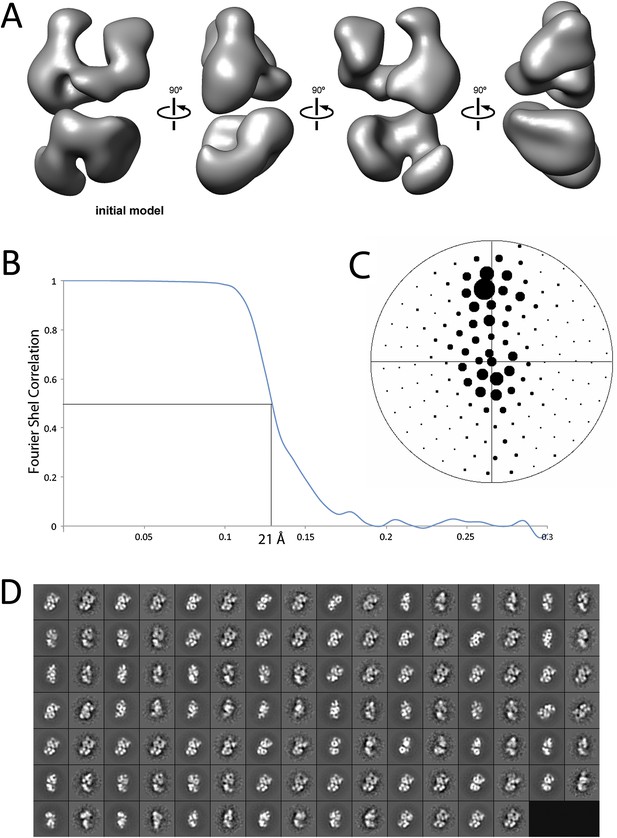
Refinement and final statistics for the reconstruction of the human PRC2-AEBP2 complex.
(A) 3D reconstruction of the ab initio model used for iterative projection-matching (corresponds to reconstruction 4 in Figure 2C). (B) Final resolution was estimated as 21 Å using the Fourier shell correlation criterion with a cutoff of 0.5. (C) Euler distribution plot of the PRC2-AEBP2 particles, where the size of the circle corresponds to the relative number of views included for that projection. (D) Comparison between re-projection of the final model (even numbers) and reference free 2D class averages (odd numbers). The projections were generated at 12° increments around the PRC2 long axis, including out-of-plane tilting going up to 30°.
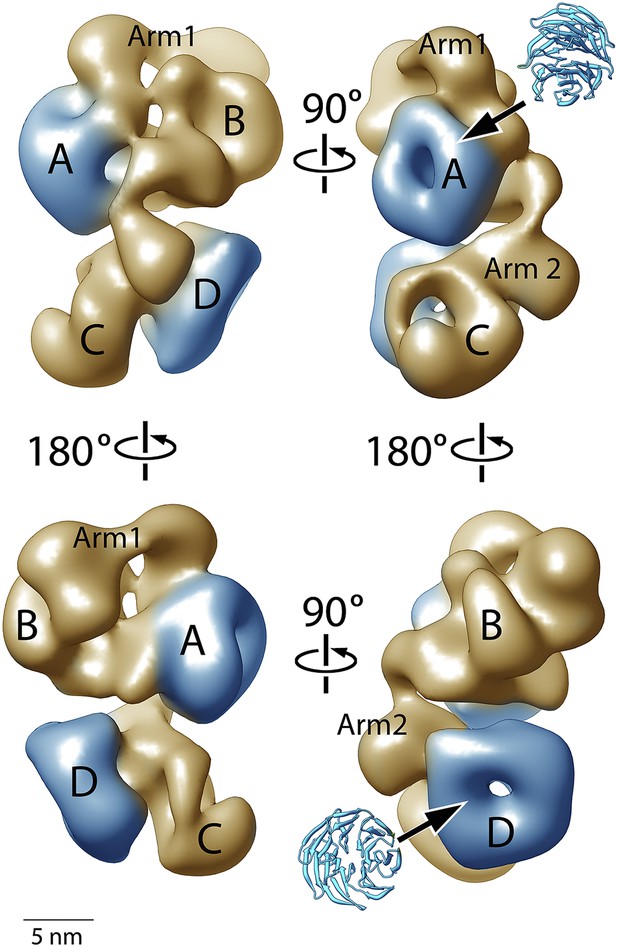
Structure of the human PRC2-AEBP2 complex.
(A) The PRC2-AEBP2 complex consists of 4 different lobes, each about 55 Å in diameter (A, B, C, D), interconnected by two narrow arms (Arm 1, Arm 2). The two WD40 domains of EED and RbAp48 (indicated in blue) are located at opposite ends.
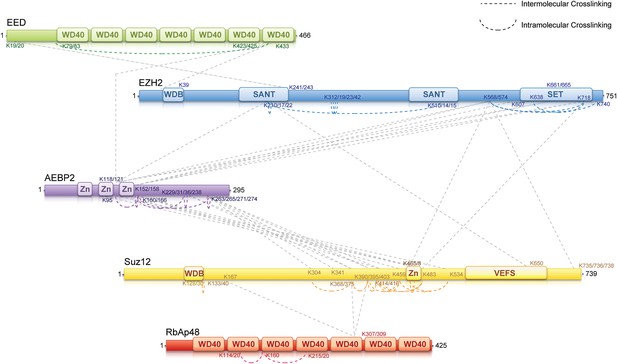
MS-coupled cross-linking analysis of the PRC2-AEBP2 complex.
Cross-link map for PRC2 in complex with AEBP2. Observed inter-molecular cross-links (straight dashed lines) are colored in grey. Intra-molecular cross-links are color coded by the respective PRC2-AEBP2 subunit (See also Supplementary file 1).
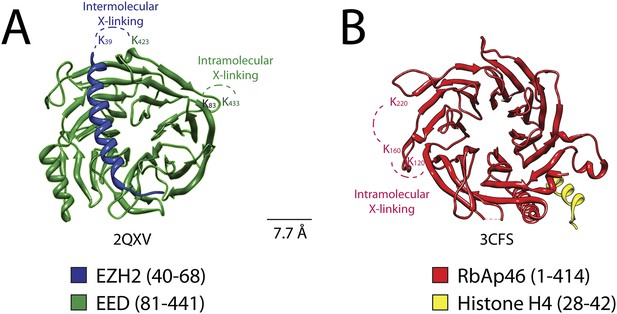
Isotopic crosslinking within the WD40 regions of EED and RbAp48.
(A) Crystal structure of EED in complex with Ezh2 (PDB: 2QXV). EED is colored in green and Ezh2 in blue. Both intermolecular and intra-molecular crosslinking data fit very well with the inter-residue distance in the structure. Scale bar of 7.7 Å represent the length of DSSG molecule used for the cross-linking experiment. (B) Structure of the subunit RbAp46 (PDB: 3CFS; 89% identity to RbAp48). Distance between residues K120 and K160 (∼5 Å) is within the range of the cross-linker. The larger distance observed between cross-linked residues K160 and K220 (∼10 Å) could be justified if the length of both lysine residues is also included in the calculation.
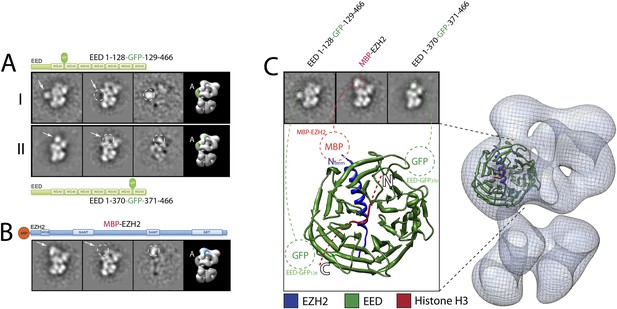
Localization and arrangement of the subunit EED within the PRC2 electron density map.
Three different internal GFP tags (EED-EGFP128, EED-EGFP370 and MBP-Ezh2) were used to determine the correct position of the EED-Ezh2 crystal structure (PDB: 2QXV) within the EM reconstruction. (A and B) For each study, a cartoon indicates the position of the tag within the protein. MBP tags are indicated in red, GFP tags in green. The far left panel represents the reference-free 2D class obtained from the labeled sample. The middle left panel shows the corresponding class for the unlabeled sample. The middle right panel was calculated by subtracting the labeled reference-free class from the unlabeled average (labeled and unlabeled). Only differences with a standard deviation greater than 3, are considered significant. The far right panel includes a representative 3D view of the complex, the localized density color-coded and the assigned Lobe indicated. (C) Top panel: 2D class averages for each mutant indicates the position of the specific tag on the molecule. Bottom panel: fitting of the X-ray structure within the EM density based on the specific position of the tags. Docking of the Histone H3 based on its crystal structure in complex with the subunit EED (PDB: 3IIW) is also indicated.
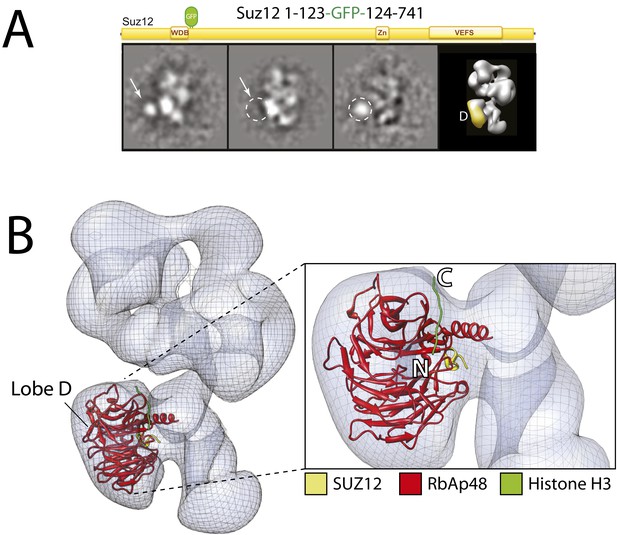
Localization of the subunit RbAp48 within the PRC2-AEBP2 electron density map.
(A) Reference-free 2D class of the labeled, unlabeled sample, difference map and 3D view of the complex with assigned localization. (B) Fitting of the Nurf55-Suz12 crystal structure within the density of Lobe D. The density has been automatically placed using the local localization algorithm implemented in the UCSF Chimera software (Pettersen et al., 2004). Docking of the Histone H3 based on its crystal structure in complex with the RbAp48 homologue subunit Nurf55 (PDB: 2YBA) is also indicated.
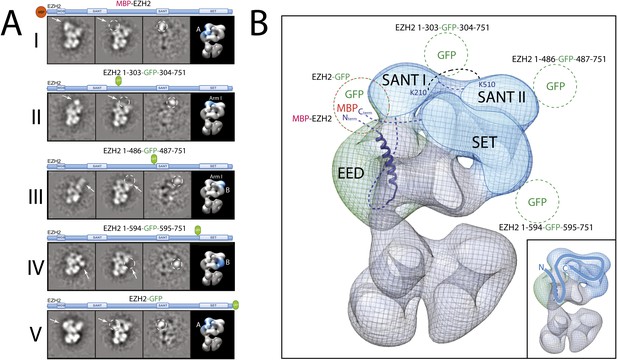
Localization and arrangement of the subunit EZH2 within the PRC2-AEBP2 electron density map.
(A) Reference-free 2D class of the labeled, unlabeled sample, difference map and 3D view of the complex with assigned localization for different MBP/GFP fusion mutants of EZH2. (B) Assignment of EZH2 subunit domains to electron density map based on the specific position of the tags. The crosslink between K210 and K510 is shown. The EZH2 protein chain path within the PRC2-AEBP2 complex is indicated in the inset (See also Figure 9—figure supplement 1).
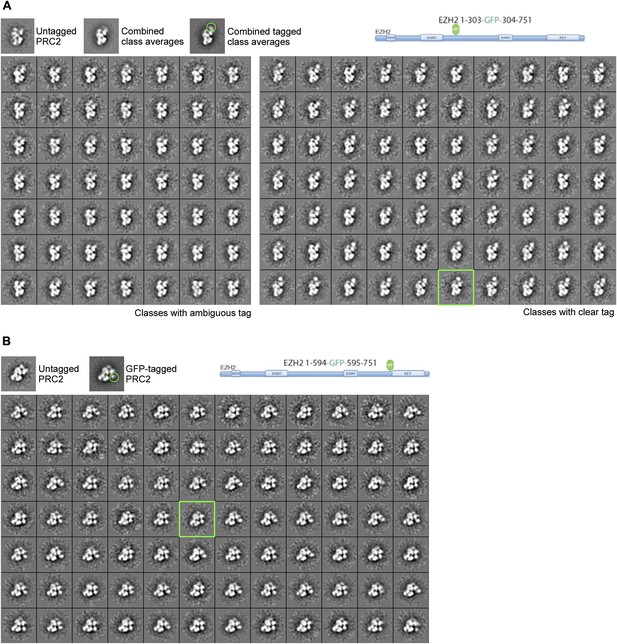
Subclassification of particles from labeled complexes illustrate the flexibility of the tag.
(A) Subclassification of the canonical view for the EZH2 1-594-GFP-595-751 complex showed all the classes to contain an extra tag density. The class average selected for Figure 9AIV is indicated with a green box. Notice that the GFP density is clearer in any of the subclasses than in the overall average (top right panel, green circle) (See also Movie 1). (B) Subclassification of a well-represented, canonical view of the EZH2 1-303-GFP-304-751 labeled complex shows some subclasses without clear extra density (left panels) and other with clear, but flexible tag (right panel). The class shown in Figure 9AII is indicated with the green box.
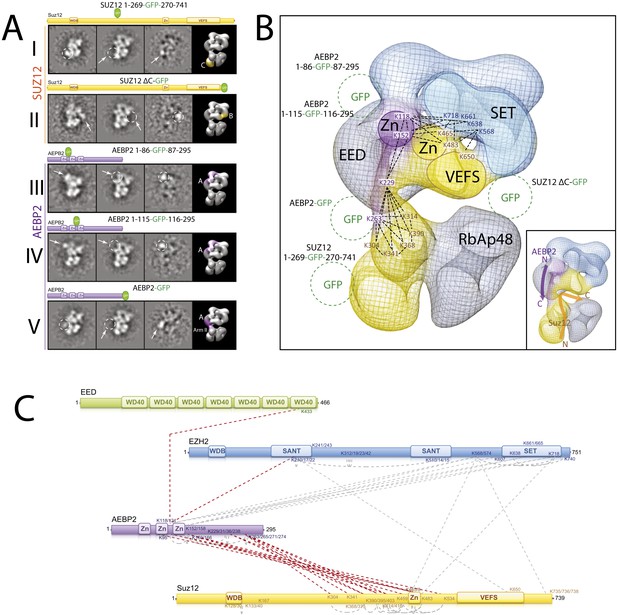
Localization and Arrangement of the subunits Suz12 and AEBP2 in the PRC2-AEBP2 electron-density map.
(A) Reference-free 2D class of the labeled, unlable sample, difference map and 3D view of the complex with assigned localization for different GFP fusion mutants of Suz12 and AEBP2. (B) Assignment of Suz12 and AEBP2 subunit domains to electron density map based on the specific position of the tags. Suz12 and AEBP2 protein chains paths within the PRC2-AEBP2 complex are indicated in the inset. Aminoacidic numbers indicate cross-linked residues in the contest of the electron density map. (C) Isotopic cross-linking of PRC2 complex. Connections in red indicate the cross-linked aminoacids referred in the text.
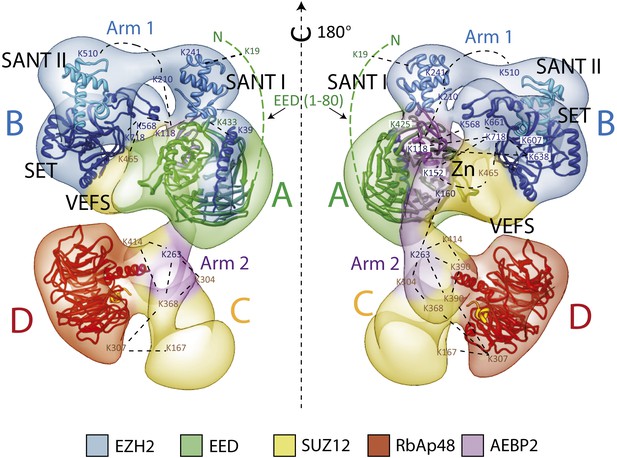
Overall Architecture of the PRC2 Complex.
Model of the Prc2 complex from a combination of biochemical data, mass spectrometry, and structural biology data. Docking of crystal structure for EED and RbAp48 WD40s (PDB: 2QXV; 2YB8) are indicated respectively in green and red. Docking of crystal structures of homologue SANT, SET and Zn finger domains (PDB: 3HM5, 3H6L and 2VY5) are shown in blue and purple. Approximate positions of crosslinking sites are also indicated. See also Movie 2.
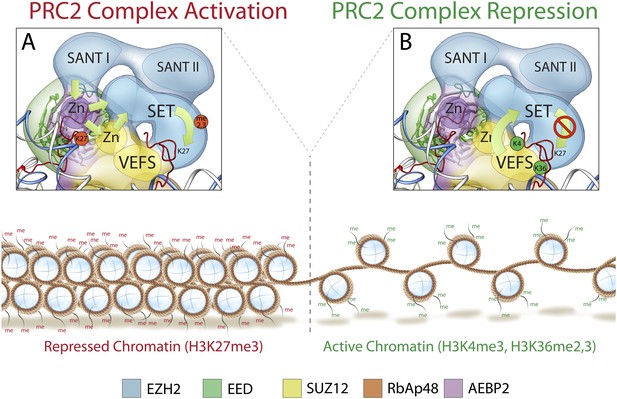
Mechanism and allosteric regulation of PRC2 during gene silencing.
(A) At loci of compact and repressed chromatin, H3K27-me3 marks are recognized by EED. This binding is signaled via the SANT domains to the SET domain increasing the methyl-transferase activity of Ezh2, strengthening the chromatin compaction. (B) At loci of open and actively transcribed chromatin, H3K4me3 and H3K36me2,3 are recognized by the VEFS domain of Suz12 and transferred to Ezh2, with an allosteric regulation that blocks Ezh2's enzymatic activity.
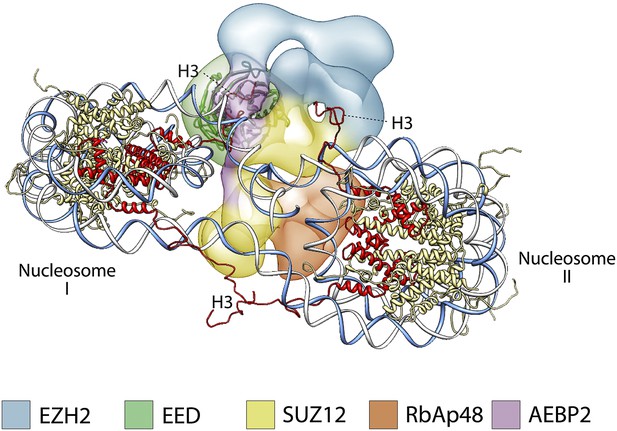
A proposed possible model for the binding of the PRC2-AEBP2 complex to a di-nucleosome.
https://doi.org/10.7554/eLife.00005.019Videos
Movement of GFP tag.
https://doi.org/10.7554/eLife.00005.0133D reconstruction of the human PRC2-AEBP2 complex.
Docking of crystal structure for EED and RbAp48 WD40s (PDB: 2QXV; 2YB8) are indicated respectively in green and red. Docking of crystal structures of homologue SANT, SET and Zn finger domains (PDB: 3HM5, 3H6L and 2VY5) are shown in blue and purple.
-
Movie 2—source code 1
Overall architecture of the PRC2 complex.
- https://doi.org/10.7554/eLife.00005.017
Additional files
-
Supplementary file 1
Inter- and intramolecular crosslinking.
- https://doi.org/10.7554/eLife.00005.020





