The unfolded protein response in fission yeast modulates stability of select mRNAs to maintain protein homeostasis
Figures
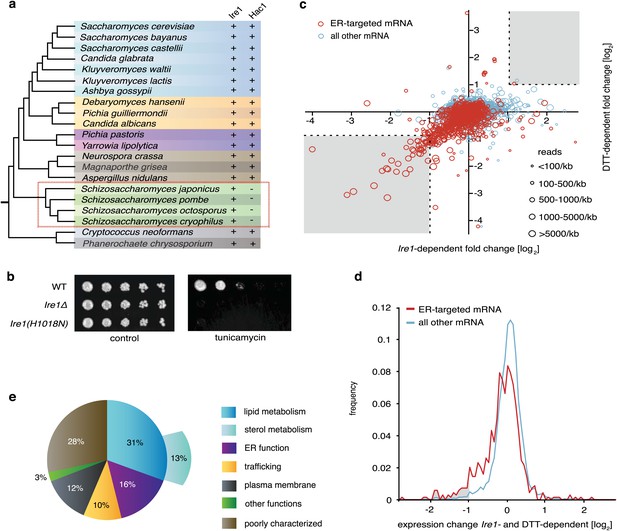
The UPR in fission selectively down-regulates ER-targeted mRNAs.
(a) Phylogenetic tree showing the components of the UPR in yeasts. The presence of recognizable orthologs of Ire1 and Hac1 is indicated. (b) Viability assay by serial dilution of wild type, Ire1Δ and Ire1(H1018N) cells spotted on solid media with or without 0.03 µg/ml of the ER stress inducer tunicamycin (Tm). Plates were photographed after 3 day of growth at 30°C. (c) Strand-specific polyA+ enriched mRNA-Seq analysis of annotated ORFs. The plot indicates the fold change (log2) of transcript abundance in DTT-stressed Ire1Δ cells (2 mM DTT, 1 hr) compared to DTT-stressed wild type cells (2 mM DTT, 1 hr) in the x-axis, and transcript abundance in unstressed wild type cells compared to DTT-stressed wild type cells (2 mM DTT, 1 hr) in the y-axis. Symbol sizes indicate abundance classes for each mRNA (reads per kilobase). Transcripts encoding proteins with a signal sequence or transmembrane segment are colored red, all other transcripts are colored blue (Figure 1—source data). (d) DTT-dependent and Ire1-dependent expression changes of transcripts displaying a signal sequence or a transmembrane domain. The skew of the left tail of the distribution indicates an enrichment (p<1×10−20) of down-regulated mRNAs. Coloring is as in Figure 1d. (e) Distribution of gene-ontology (GO) annotations for Ire1-dependent down-regulated mRNAs. Percentages indicate genes within a particular GO category in relation to the total number of genes that have a GO annotation (N=39) (see Figure 1—figure supplement 3 for annotated list of genes).
-
Figure 1—source data 1
Gene expression and fold change
- https://doi.org/10.7554/eLife.00048.004
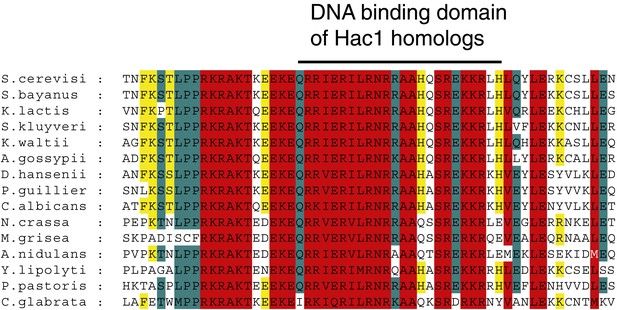
Alignment of DNA binding domain of Hac1 (bZIP) homologues in different yeast species.
https://doi.org/10.7554/eLife.00048.005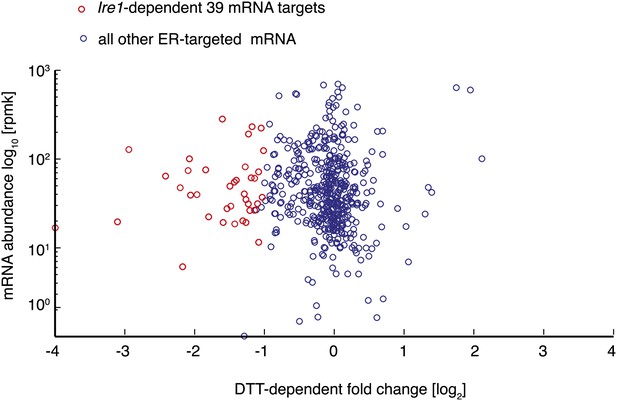
Plot depicting ER-targeted mRNAs abundance [log10] (reads per million) versus DTT-dependent expression changes [log2] for wild type cells.
https://doi.org/10.7554/eLife.00048.006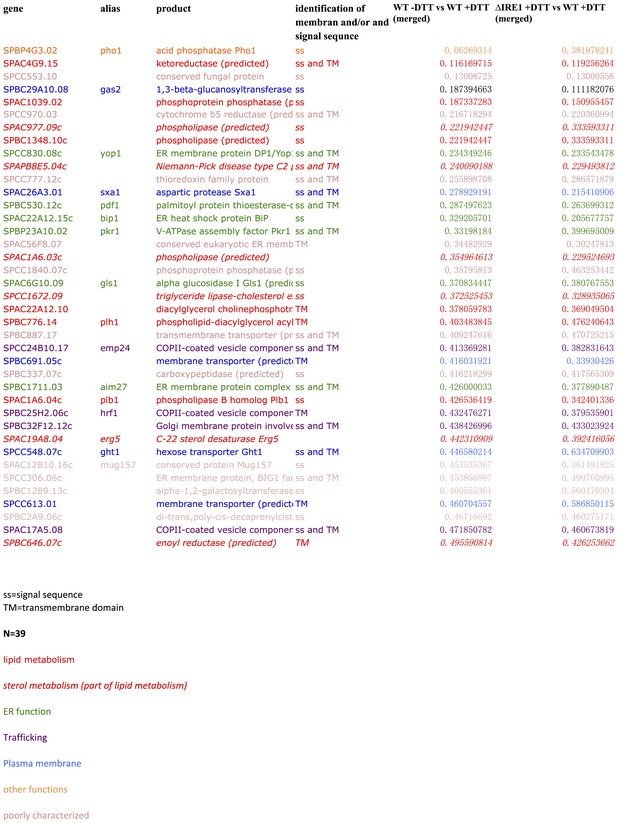
Ontology of genes down-regulated more than twofold Ire1-and DTT-dependent.
https://doi.org/10.7554/eLife.00048.007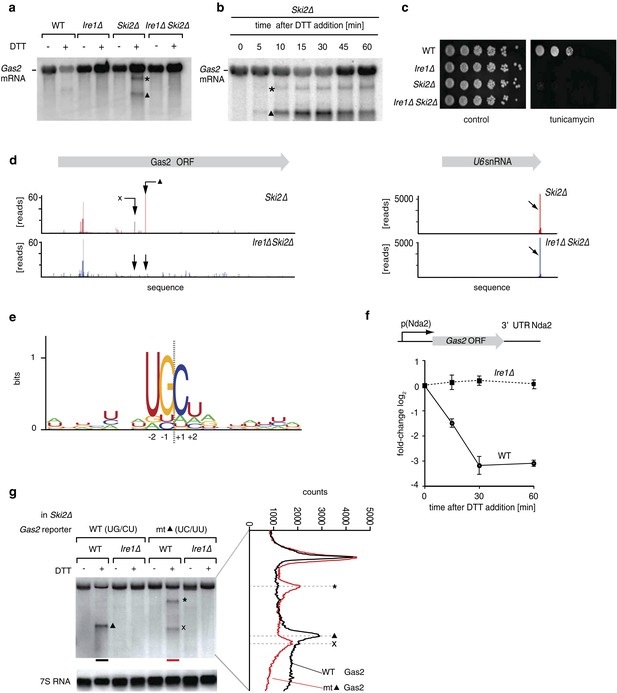
Ire1 cleaves down-regulated mRNAs at specific sequences.
(a) Northern blot of total RNA extracted from wild type, Ire1Δ, Ski2Δ and double mutant ER stressed Ire1Δ Ski2Δ cells (2 mM DTT, 1 hr). A probe complementary to the 5′ UTR of Gas2 was used to detect cleavage products. The triangle and asterisk indicate two different mRNA cleavage products. (b) Northern blot of total RNA extracted from ER-stressed Ski2Δ cells (2 mM DTT). (c) Viability assay by serial dilution of wild type, Ire1Δ, Ski2Δ and Ire1Δ Ski2Δ cells spotted on solid media with or without ER stress as in Figure 1b. (d) RNA-sequence read density map of the Gas2 locus derived from 3′ end deep-sequencing data. Library was generated by ligating a DNA-linker using tRNA ligase to 3′ end mRNAs with 2′,3′-cyclic phosphates in Ski2Δ and Ire1Δ Ski2Δ ER-stressed cells (2 mM DTT, 30 min). The arrows indicate two Ire1-depedent cleavage sites. The U6 snRNA locus was used as a positive control (Figure 2—source data). (e) Ire1 RNA sequence recognition motifs generated by deep-sequencing analysis of tRNA ligase-generated RNA libraries of 39 mRNA targets down-regulated twofold or more in an Ire1-dependent manner. The resulting position weight matrices are illustrated as a logo. The dotted line indicates the cleavage site. (f) Real-time qPCR of a chromosomally integrated reporter containing the coding sequence of Gas2 under the control of the Nda2 (tubulin) promoter and including the UTRs of Nda2. A time course after DTT addition (2 mM) is shown. Endogenous Nda2 was used as a normalization control. Error bars: standard deviation. (g) Northern blot analysis of total RNA extracted from Ski2Δ and Ire1Δ Ski2Δ cells carrying a mutant version of the reporter indicated in (f) were the putative Ire1 cleavage site (▲, UG\CU→ UC\UU) was mutated. Note that the band labeled × migrates distinctly faster, as shown by scan on the right.
-
Figure 2—source data 1
2′,3′ cyclic-phosphate 3′ end mapping
- https://doi.org/10.7554/eLife.00048.009
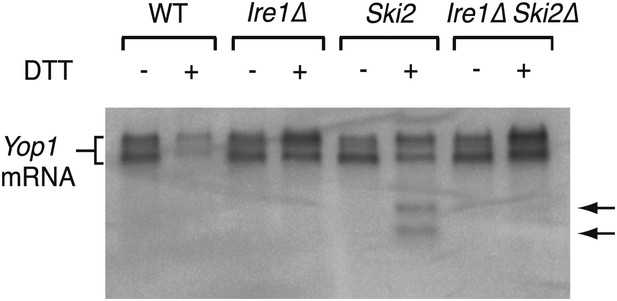
Northern blot of total RNA isolated from wild type, Ire1Δ, Ski2Δ and double mutant Ire1Δ Ski2Δ ER stressed cells (2 mM DTT, 1 hr).
A probe complementary to the 5′ UTR of Yop1 was used to detect cleavage products. The arrows indicate Ire1-dependent mRNA cleavage products.
Comparison of ER stress-dependent down-regulation of endogenous Gas2 mRNA (2 mM DTT, 1 hr: deep-sequencing) and reporter Gas2 mRNA (from qPCR: 2 mM DTT, 1 hr; see also Figure 2f).
https://doi.org/10.7554/eLife.00048.011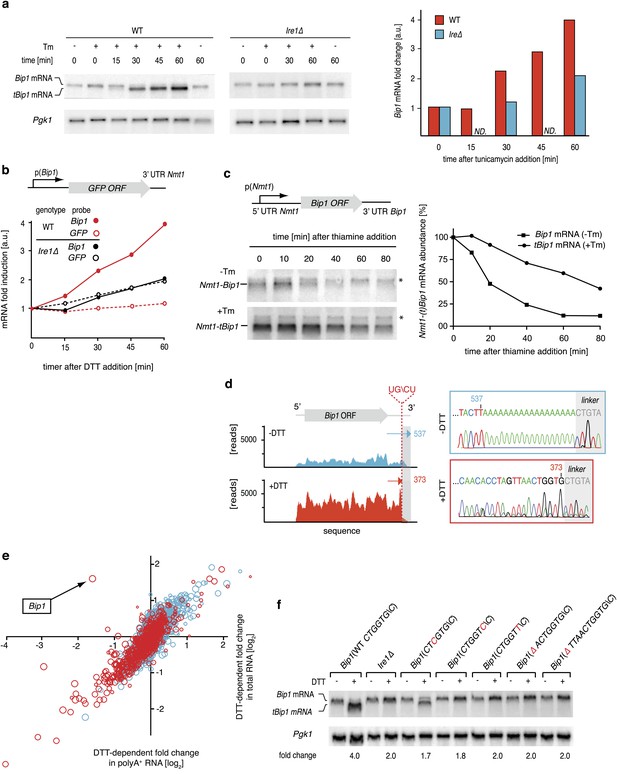
Ire1 truncates Bip1 mRNA within the 3′ UTR.
(a) Northern blot analysis of total RNA extracted from wild type and Ire1Δ cells untreated or treated with tunicamycin (1 µg/ml), and hybridized with a probe complementary to the ORF of Bip1 mRNA. Right panel: quantitation normalized to Pgk1 mRNA. (b) The abundance of a GFP mRNA driven by the Bip1 promoter (black) compared to endogenous Bip1 mRNA was determined as a time course after DTT (2 mM) addition by quantitative Northern blotting. (c) Wild type cells bearing a construct encoding the Nmt1 5′ UTR, Bip1 ORF and Bip1 3′ UTR driven by the Nmt1 promoter were pre-treated with tunicamycin (0.25 µg/ml, 1 hr). At different time points after thiamine (15 µM) addition (to effect transcriptional shut-off of the Nmt1 promoter), RNA was extracted and analyzed by Northern hybridization. Blots were probed for the Nmt1 5′ UTR. Nmt1-Bip1 mRNA and Nmt-tBip1 mRNA were quantitated and normalized to the unspecific band (asterisk). (d) RNA-sequence read density map of the Bip1 locus derived from mRNA-enriched (ribosome depleted) RNA in wild type cells untreated or treated with DTT (2 mM DTT, 1 hr; left panels). Data are representative of one of two biological replicates. Single nucleotide resolution of the 3′ terminus of Bip1 mRNA determined by 3′ RACE (right panels). (e) Mutational analysis of the Bip1 mRNA cleavage site by Northern blotting. Total RNA was extracted from wild type, Ire1Δ or cells carrying mutations in the Bip1 3′ UTR mRNA. Cells were treated with 2 mM DTT, 1 hr or left untreated as indicated. The fold-changes indicate Bip1 mRNA abundance relative to that of Pgk1 mRNA. (f) Strand-specific, mRNA enriched (after removal of ribosomal RNA) deep-sequence analysis of annotated ORFs (y-axis) compared to strand-specific polyA+ enriched mRNA deep-sequence analysis of annotated ORFs (x-axis) (see Figure 1—source data). The plot indicates the ratio of transcript abundance in unstressed vs DTT-stressed (2 mM DTT, 1 hr) wild type cells. Symbol sizes and colors are as described in Figure 1c.
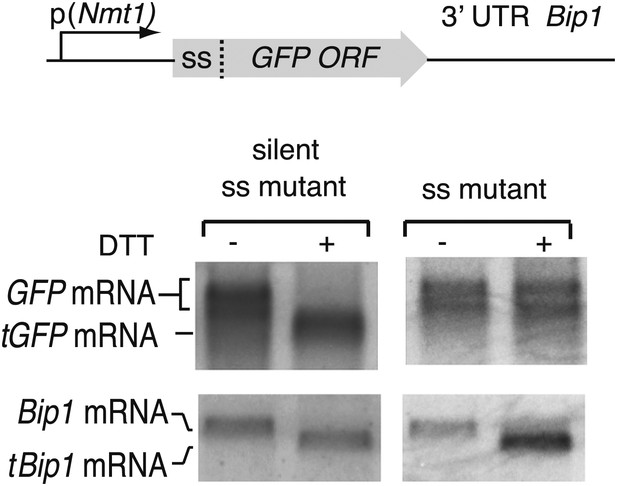
Northern blot analysis of wild type cells bearing a construct expressing a fusion protein of GFP preceded by the Bip1 signal sequence.
The constructs includes the Bip1 3' UTR. Expression was driven by the Nmt1 promoter. Cells were untreated or treated with DTT (2 mM DTT, 1 hr) as indicated. To abolish targeting of the mRNA to the ER, three hydrophobic leucine residues in the signal sequence were replaced by three charged arginine residues. MKKFQLFSILSYFVALFLLPMAFA (WT) to MKKFQRFSIRSYFVARFLLPMAFA. Silent mutations were created by changing one nucleotide in each three leucine residues (above) without changing the amino acid.
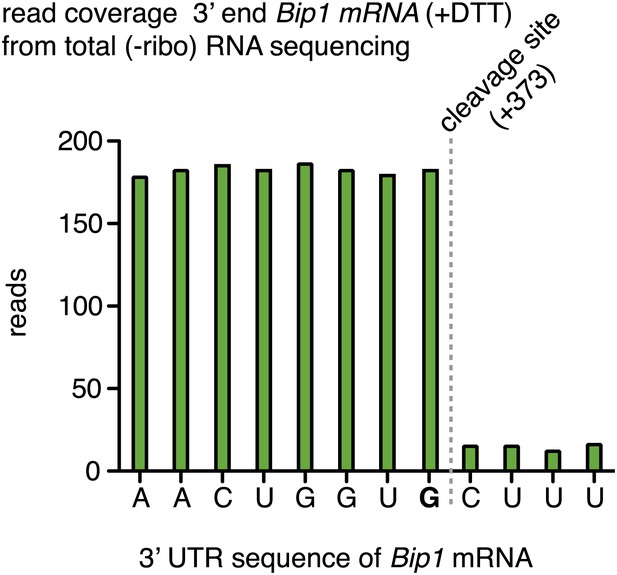
Sequencing read coverage of the 3′; end nucleotide positions in tBip1 mRNA from derived from mRNA enriched by subtractive hybridization against rRNA (Ribominus kit, Invitrogen kit).
Cells were treated with DTT (2 mM, 1 hr).
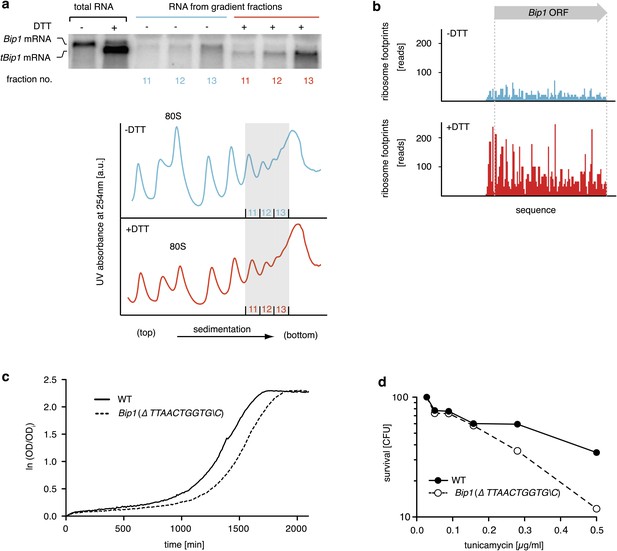
tBip1 mRNA is translated and is important for fitness during ER stress.
(a) Northern blot analysis of the distribution of total or tBip1 mRNA in polyribosomes from extracts of unstressed or ER-stressed (2 mM DTT, 1 hr) cells. Fractions 11, 12, 13 of the sucrose gradients (lower panels) were analyzed by Northern blotting (upper panels). (b) Ribosome footprints (as described in Materials and methods) of Bip1 mRNA in unstressed or ER stressed cells. The region that depicts ribosome density preceding the Bip1 ORF is shown enlarged in Figure 4—figure supplement 1 (Figure 4—source data 1). (c) Cell growth of wild type cells and cells carrying a deletion of Bip1 mRNA cleavage sites (Bip1(Δ TTAACTGGTGC)). Cells were treated with tunicamycin (0.5 µg/ml) for 3 hr and then recovered from ER stress by washing out the drug and re-seeding in warm fresh media. Optical density (OD) at 660 nm was measured immediately afterward in 10 min intervals. (d) Viability assay of the same cells as in (c). The percentage of viable cells was determined by counting the number of colony-forming units (CFU) after growth for 3 hr at varying tunicamycin concentration.
-
Figure 4—source data 1
Ribosome footprint reads for Bip1
- https://doi.org/10.7554/eLife.00048.016
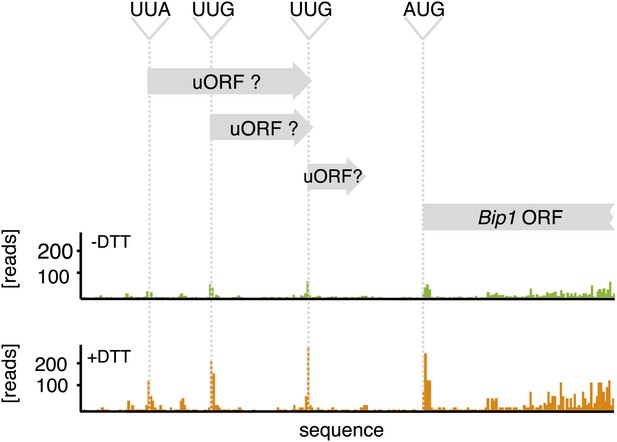
Zoomed-in ribosome occupancy profile of Bip1 mRNA around the start AUG codon of wild type cells. Putative non-canonical uORFs are highlighted in both Bip1 and tBip1 mRNA derived from untreated and DTT-treated (2 mM, 1 hr) cells.
https://doi.org/10.7554/eLife.00048.017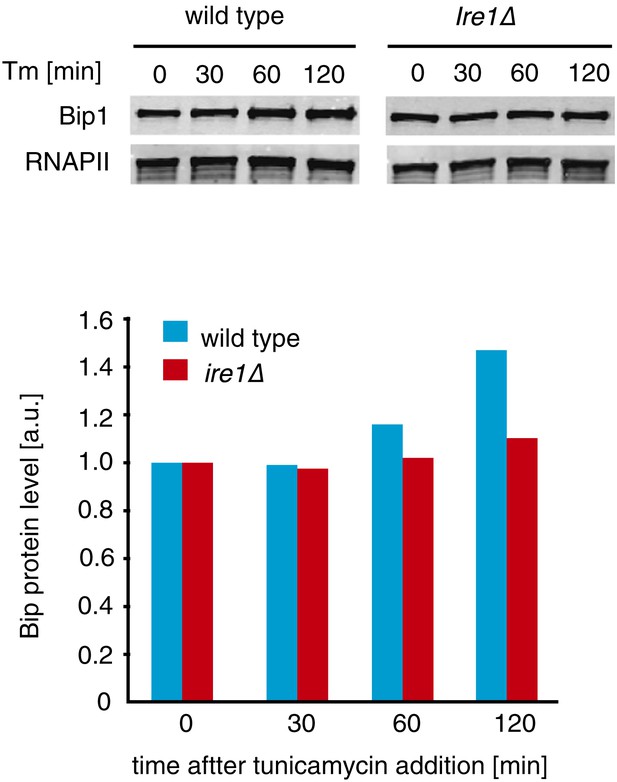
Upper panel: Western blot of Bip1 and, as a loading control, RNA polymerase II CTD repeat (RNAPII). Wild type and Ire1Δ cells were treated with tunicamycin 0.5 µg/ml and samples were taken at indicated time points. Lower panel: Quantification of Western blotting with values normalized to RNAPII.
https://doi.org/10.7554/eLife.00048.018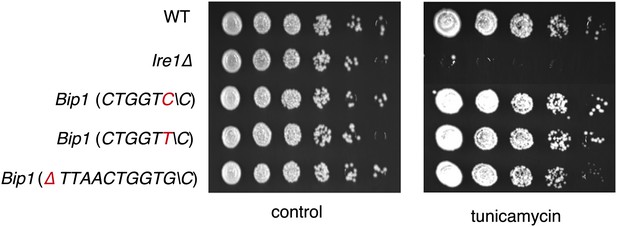
Viability assay by serial dilution of wild type, Ire1Δ and different Bip1 mRNA cleavage mutants spotted on solid media with or without 0.03 µg/ml of the ER stress inducer tunicamycin. Plates were photographed after 3 day of growth at 30°C.
https://doi.org/10.7554/eLife.00048.019Tables
Strains and plasmids used in this study
| Strain | Description | |
| yPK001 | WT | |
| yPK002 | Ire1Δ | |
| yPK003 | Ire1(H1018N) | |
| yPK004 | Ski2Δ | |
| yPK005 | Ski2Δ Ire1Δ | |
| yPK006 | Bip1 (CTCGTG\C) | |
| yPK007 | Bip1 (CTGGTC\C) | |
| yPK008 | Bip1 (CTGGTT\C) | |
| yPK009 | Bip1 (ΔACTGGTG\C) | |
| yPK010 | Bip1 (ΔTTAACTGGTG\C) | |
| Plasmid | Description | Marker |
| pPK001 | pJK148::pNda2::Gas2::Nda2 3′UTR | Leu1 |
| pPK002 | pJK148::pBip1::GFP::Nmt1 3′UTR | Leu1 |
| pPK003 | pJK148::pNmt1+5′UTR::Bip1(ORF)::Bip1 3′UTR | Leu1 |
| pPK004 | pJK148::pNmt1::ss(Bip1)::GFP::Bip1 3′UTR | Leu1 |
Ribosomes footprints of mRNAs encoding protein predicted to enter the ER
| All ER-targeted mRNAs ribosome occupancies (N=1014 mRNAs) | rpkm | Ration normalized to WT-DTT | % | |
| WT (-DTT) | 83,540 | 1 | 100 | |
| WT(+DTT) | 71,996 | 0.8618147 | 86.18146995 | |
| Ire1∆( +DTT) | 90,166 | 1.079315298 | 107.9315298 |
-
rpkm: reads per kilo-base of transcript per million reads.





