RNA-programmed genome editing in human cells
Figures
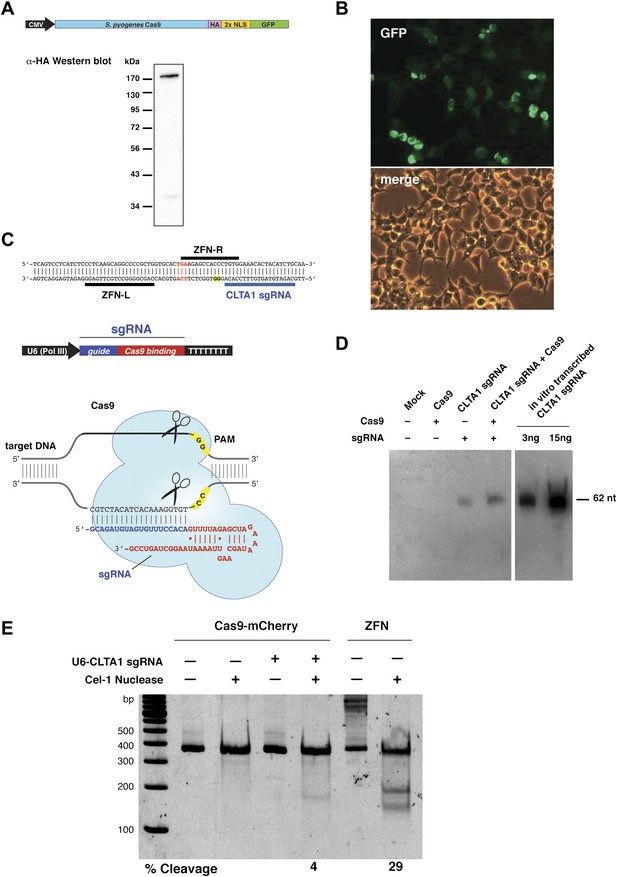
Co-expression of Cas9 and guide RNA in human cells generates double-strand DNA breaks at the target locus.
(A) Top: schematic diagram of the Cas9-HA-NLS-GFP expression construct. Bottom: lysate from HEK293T cells transfected with the Cas9 expression plasmid was analyzed by Western blotting using an anti-HA antibody. (B) Fluorescence microscopy of HEK293T cells expressing Cas9-HA-NLS-GFP. (C) Blue line indicates the sequence used for the guide segment of CLTA1 sgRNA. Top: schematic diagram of the sgRNA target site in exon 7 of the human CLTA gene. The target sequence that hybridizes to the guide segment of CLTA1 sgRNA is indicated by the blue line. The GG nucleotide protospacer adjacent motif (PAM) is highlighted in yellow. Black lines denote the DNA binding regions of the control ZFN protein. The translation stop codon of the CLTA open reading frame is highlighted in red for reference. Middle: schematic diagram of the sgRNA expression construct. The RNA is expressed under the control of the U6 Pol III promoter and a poly(T) tract that serves as a Pol III transcriptional terminator signal. Bottom: sgRNA-guided cleavage of target DNA by Cas9. The sgRNA consists of a 20-nt 5′-terminal guide segment (blue) followed by a 42-nt stem-loop structure required for Cas9 binding (red). Cas9-mediated cleavage of the two target DNA strands occurs upon unwinding of the target DNA and formation of a duplex between the guide segment of the sgRNA and the target DNA. This is dependent on the presence of a GG dinucleotide PAM downstream of the target sequence in the target DNA. Note that the target sequence is inverted relative to the upper diagram. (D) Northern blot analysis of sgRNA expression in HEK239T cells. (E) Surveyor nuclease assay of genomic DNA isolated from HEK293T cells expressing Cas9 and/or CLTA sgRNA. A ZFN construct previously used to target the CLTA locus (Doyon et al., 2011) was used as a positive control for detecting DSB-induced DNA repair by non-homologous end joining.
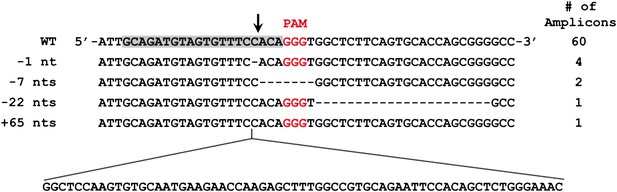
Mutated alleles of the CLTA gene in HEK293T cells as a result of Cas9-induced NHEJ.
Four types of indels were identified by Sanger sequencing in 68 clonal amplicons. The protospacer sequence is highlighted in grey with the putative cleavage site indicated by the arrow. Dashes represent deleted bases.
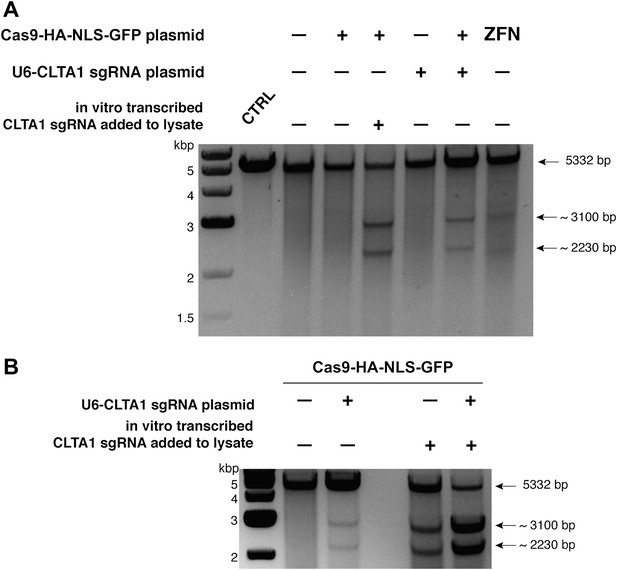
Cell lysates contain active Cas9:sgRNA and support site-specific DNA cleavage.
(A) Lysates from cells transfected with the plasmid(s) indicated at left were incubated with plasmid DNA containing a PAM and the target sequence complementary to the CLTA1 sgRNA; where indicated, the reaction was supplemented with 10 pmol of in vitro transcribed CLTA1 sgRNA; secondary cleavage with XhoI generated fragments of ∼2230 and ∼3100 bp fragments indicative of Cas9-mediated cleavage. A control reaction using lysate from cells transfected with a ZFN expression construct shows fragments of slightly different size reflecting the offset of the ZFN target site relative to the CLTA1 target site. (B) Lysates from cells transfected with Cas9-HA-NLS-GFP expression plasmid and, where indicated, the CLTA1 sgRNA expression plasmid, were incubated with target plasmid DNA as in (A) in the absence or presence of in vitro-transcribed CLTA1 sgRNA.
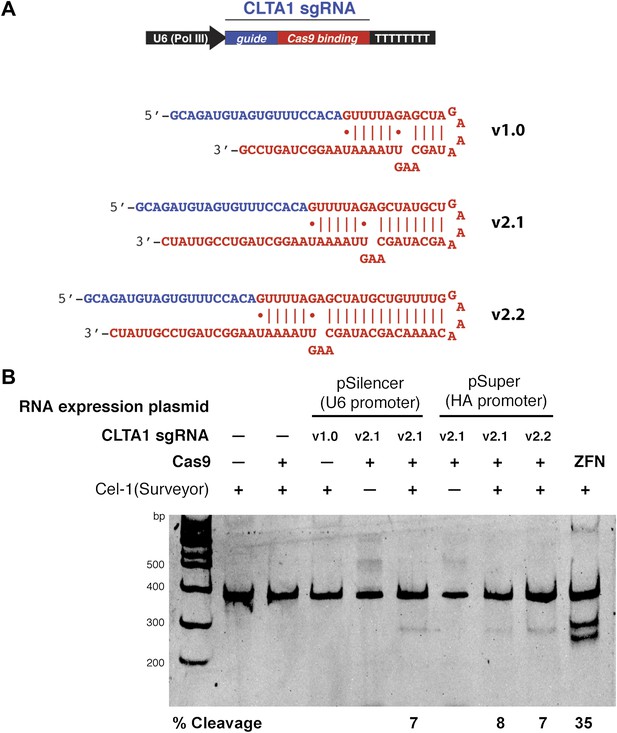
3′ extension of sgRNA constructs enhances site-specific NHEJ-mediated mutagenesis.
(A) The construct for CLTA1 sgRNA expression (top) was designed to generate transcripts containing the original Cas9-binding sequence v1.0 (Jinek et al., 2012), or sequences extended by 4 base pairs (v2.1) or 10 base pairs (v2.2). (B) Surveyor nuclease assay of genomic DNA isolated from HEK293T cells expressing Cas9 and/or CLTA sgRNA v1.0, v2.1 or v2.2. A ZFN construct previously used to target the CLTA locus (Doyon et al., 2011) was used as a positive control for detecting DSB-induced DNA repair by non-homologous end joining.
Additional files
-
Supplementary file 1
DNA and protein sequences relating to plasmid design and construction.
- https://doi.org/10.7554/eLife.00471.007





