Accurate timekeeping is controlled by a cycling activator in Arabidopsis
Figures
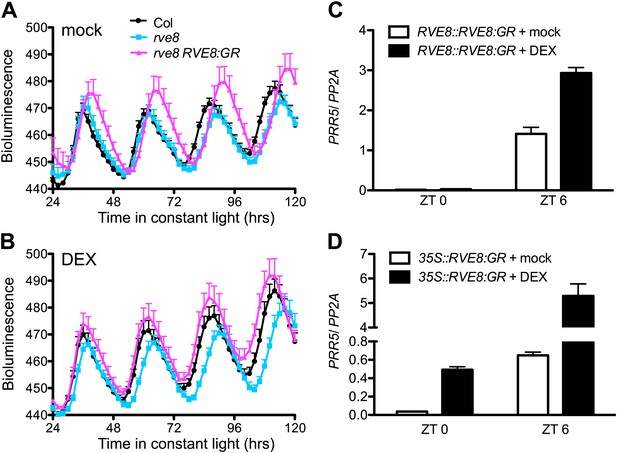
Activation of PRR5 by RVE8 induction is stronger in the afternoon.
(A) and (B) Luciferase activity in mock (A) and DEX-treated (B) Col, rve8-1 and rve8-1 RVE8::RVE8:GR plants transgenic for the CCR2::LUC reporter. Plants were entrained in 12:12 light/dark (LD) cycles for 6 days and then sprayed with 30 µM DEX or 0.05% ethanol (mock treatment) plus luciferin before release to constant red light (30 µEi) for imaging of bioluminescence. Mean + SEM from 17 to 25 plants are represented. (C) and (D) Transcript levels of PRR5 in response to induction of RVE8 activity in rve8-1 RVE8::RVE8:GR (C) or rve8-1 35S::RVE8:GR (D) at different time of day. 30 µM DEX or 0.05% ethanol (mock) was applied at the times indicated and the plants were harvested 2 hr later. Expression levels were quantified by qRT-PCR and normalized to PP2A. Mean ± SEM from three biological replicates are represented.
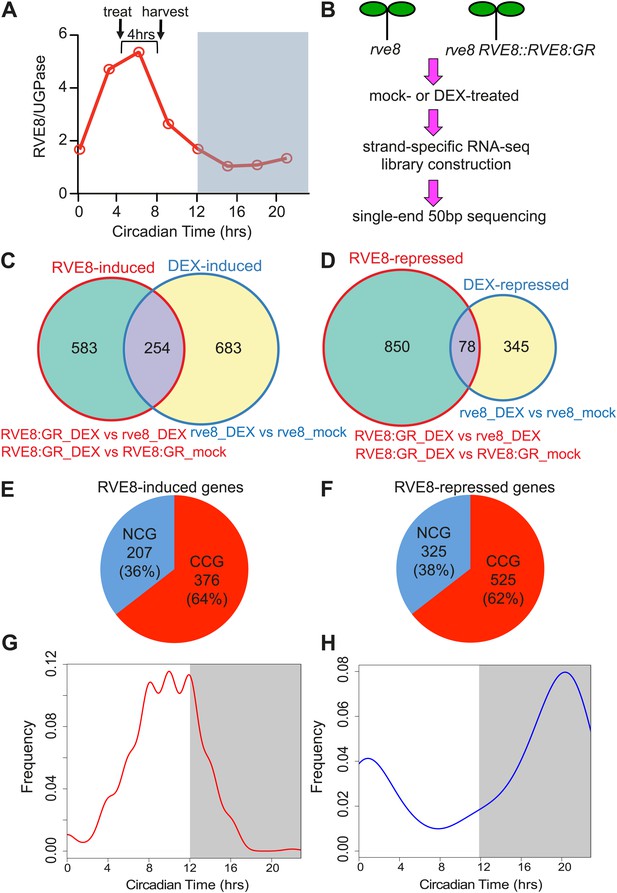
Identification of RVE8 targets by RNA-seq. RNA-seq experimental design and data analysis.
(A) Relative timing of RVE8 induction and RVE8 protein abundance during a day. Adapted from Rawat et al. (2011). (B) Scheme of experimental design. (C) and (D) Weighted Venn diagrams of genes significantly responsive to RVE8 induction and/or DEX treatment. Genes up-regulated (C) or down-regulated (D) by RVE8 and/or DEX. Differentially expressed genes were identified using edgeR (Robinson et al., 2010) with an adjusted p-value <0.01 as the cutoff. Genes significantly different between ‘RVE8:GR + DEX’ and ‘RVE8:GR + mock’ or between ‘RVE8:GR + DEX’ and ‘rve8 + DEX’ are grouped into the RVE8-induced (C) or RVE8-repressed sets (D) shown in red circles. Genes significantly different between ‘rve8 + DEX’ and ‘rve8 + mock’ are grouped into the ‘DEX-induced’ (C) or ‘DEX-repressed’ (D) sets shown in blue circles. The genes uniquely induced or repressed by RVE8 (the 583 and 850 genes shown in green areas in (C) and (D), respectively) were defined as RVE8-regulated and used for further analysis. (E) and (F) The relative proportion of clock-controlled genes (CCGs) and non-clock-controlled genes (NCGs) among RVE8 targets. RVE8-induced genes (E); RVE8-repressed genes (F). (G) and (H) Circadian phase distributions of RVE8-regulated CCGs. CCGs up-regulated by RVE8 (G); CCGs down-regulated by RVE8 (H). White box: subjective day; grey box: subjective night. X-axis, 0: subjective dawn, 12: subjective dusk. Phase estimates are from previously published data (Hsu and Harmer, 2012). See also Supplementary file 1.
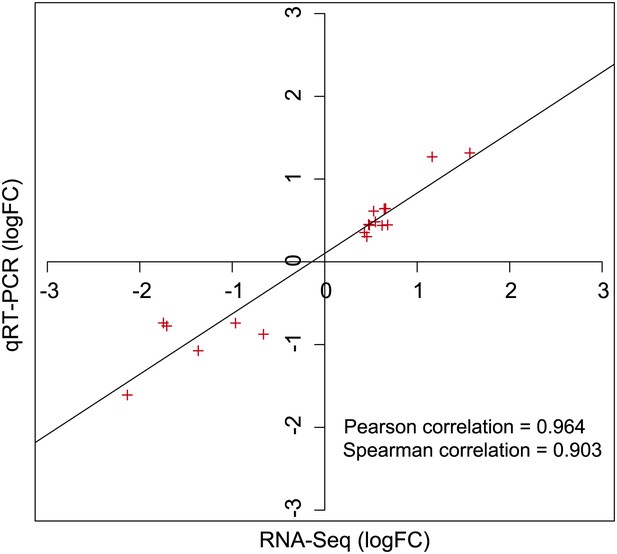
Expression levels as determined by RNA-seq and qRT-PCR are highly correlated.
Expression levels of selected genes defined as RVE8-regulated in the RNA-seq experiment were examined using qRT-PCR. The logarithm of fold change values in the RNA-seq and the qRT-PCR data were plotted along with the linear regression line to examine the correlation relationship between the two methods. Pearson and Spearman correlation tests were performed in R.
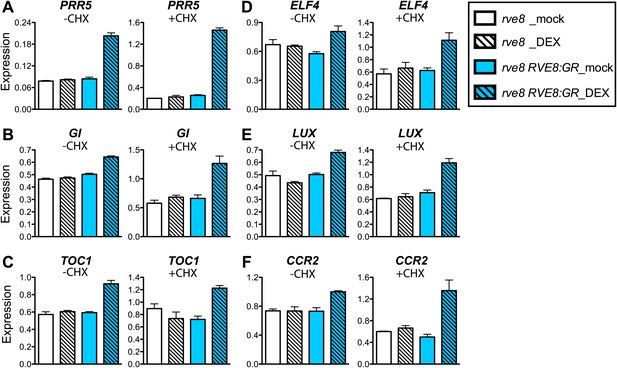
RVE8 activates evening genes directly.
(A)–(F) Transcript levels of evening genes in response to RVE8 induction in the absence or presence of cycloheximide (CHX). 7-day-old rve8-1 and rve8-1 RVE8::RVE8:GR plants were grown in light:dark (LD) cycles and mock- or DEX-treated in the absence or presence of CHX at ZT4 (4 hr after dawn) and harvested at ZT8 (8 hr after dawn). (A–E) Evening-phased clock genes. (F) Evening-phased clock output gene. Transcript levels were determined by qRT-PCR and then normalized to PP2A. Mean ± SEM from three biological replicates are represented.
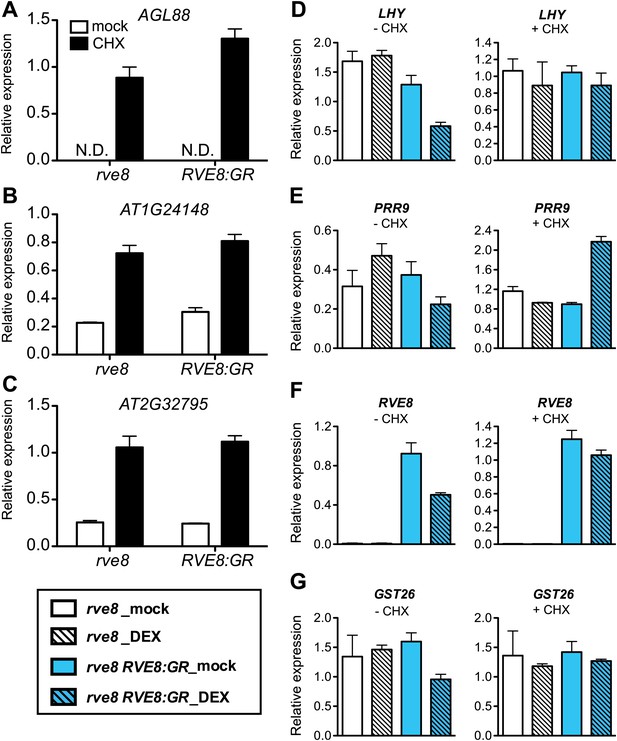
Morning-phased genes are indirectly repressed in response to RVE8 induction.
(A)–(C) Levels of transcripts controlled by nonsense-mediated mRNA decay (NMD) in response to cycloheximide (CHX) treatment. N.D.: not detectable. (D)–(G) Transcript levels of morning genes in response to RVE8 induction in the absence or presence of CHX. 7-day-old rve8-1 and rve8-1 RVE8::RVE8:GR plants were grown in light:dark (LD) cycles and mock- or DEX-treated in the absence or presence of CHX at ZT4 (4 hr after dawn) and harvested at ZT8 (8 hr after dawn). (D)–(F) Morning-phased clock genes. (G) Morning-phased clock output gene. Transcript levels were determined by qRT-PCR and then normalized to PP2A. Mean ± SEM from three biological replicates are represented.
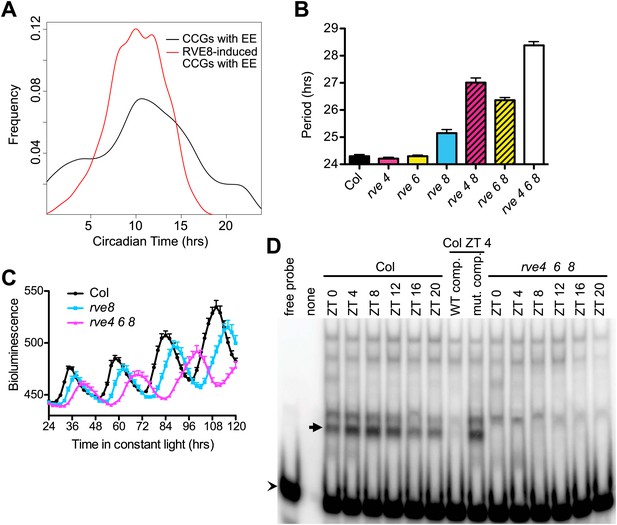
RVE8 functions through the EE.
(A) Circadian phase distributions of all EE-containing CCGs and RVE8-induced EE-containing CCGs. The RVE8-induced EE-containing CCGs are enriched for an earlier phase than that of all EE-containing CCGs. The means of the phase distribution in these two groups (10.03 for RVE8-induced EE-containing CCGs; 10.75 for all EE-containing CCGs) are significantly different (p=0.007; Student's t-test). (B) Period of CCR2::LUC activity in rve4-1, rve6-1 and rve8-1 single, double and triple mutants. Seedlings were grown in LD for 6 days and released to constant red plus blue light. Mean ± SEM from 34 to 50 plants. (C) Circadian rhythms are lengthened but still robust in rve4 rve6 rve8 mutants. Averaged bioluminescence of CCR2::LUC activity in Col, rve8-1 and rve4 rve6 rve8 triple mutants. Mean ± SEM from 20 to 25 plants. (D) An electrophoretic mobility shift (EMSA) assay with protein extracts made from Col and rve4 rve6 rve8 plants grown in LD for 11 days. Plants were harvested at the indicated times. A 50-fold molar excess of unlabeled EE (WT competitor) or mutated EE (mutant competitor) double-stranded DNA was added as indicated. Arrow: the predominant afternoon EE-binding activity, arrowhead: unbound probe. See also Figure 4—figure supplement 1.
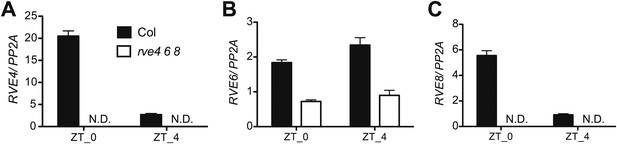
Characterization of RVE4, RVE6, and RVE8 mutant alleles.
(A)–(C) RVE4, RVE6 and RVE8 transcript levels in Col and the rve4 rve6 rve8 triple mutant. 7-day-old seedlings (about 30 plants each) were grown in 12:12 LD and harvested at ZT 0 and ZT 4. RNA was isolated and qRT-PCR was performed. RVE4 and RVE8 transcripts are not detectable (N.D.) but ∼30% of normal RVE6 transcript levels are apparent in the triple mutant. Expression levels are normalized to PP2A. Mean ± SEM from three technical replicates are presented.
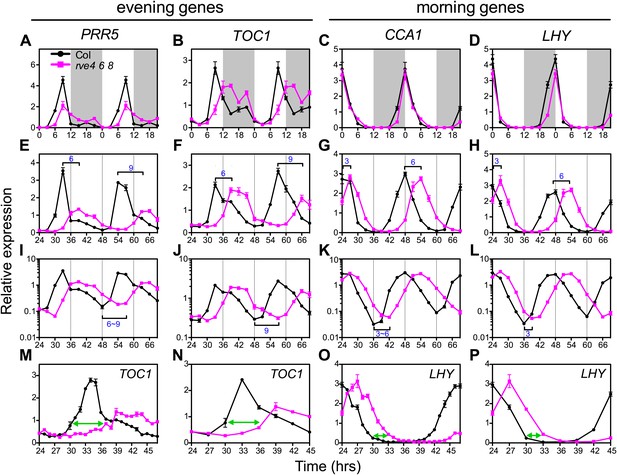
Expression of clock genes is altered in rve4 rve6 rve8 triple mutants.
(A), (B), (E), (F), (I), (J), (M) and (N) Expression of evening genes in Col and rve4 rve6 rve8. (C), (D), (G), (H), (K), (L), (O) and (P) Expression of morning genes in Col and rve4 rve6 rve8. (A)–(D) transcript levels in diurnal cycles. Seedlings were grown in LD for 7 days. White box: day, grey box: night. Data in (A–D) are double plotted to facilitate comparisons. (E)–(P) Transcript levels in LL. (E)–(H) Gene expression plotted on a linear scale. (I)–(L) The data shown in (E–H) are plotted with a log10 scale on the y-axis to better visualize differences in trough levels between the two genotypes. Horizontal brackets highlight the phase delay between Col and rve4 rve6 rve8 mutants. (M)–(P) Transcript levels derived from a 1-hr resolution time course are presented with either every time point (M and O) or every third time point (N and P) displayed. Green arrows highlight the phase difference between Col and rve4 rve6 rve8 mutants at ZT30. Transcript levels were determined by qRT-PCR and normalized to PP2A. Values represent mean ± SEM.
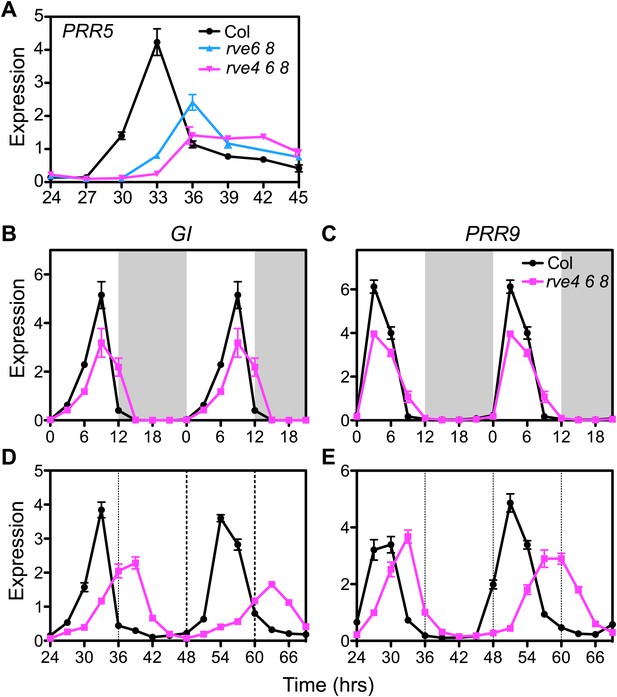
Clock gene expression in wild type and the rve4 rve6 rve8 mutant.
(A) PRR5 expression in Col, rve6 rve8 and rve4 rve6 rve8 in LL. Seedlings were grown in LD for 7 days, released to constant light, and then harvested at the indicated times after the last dark-to-light transition. (B) and (C) Expression of GI and PRR9 in diurnal cycles. Seedlings were grown in LD for 7 days. White box: day, grey box: night. Values represent mean ± SEM. Data in (B) and (C) are double plotted to facilitate comparisons. (D) and (E) Transcript levels of GI and PRR9 in LL. Seedlings were entrained in 12:12 LD for 7 days and then released to constant light at time 0. Samples were harvested at the times indicated. RNA was isolated and qRT-PCR was performed. Expression levels are normalized to PP2A. Data are presented as mean ± SEM from three technical replicates.
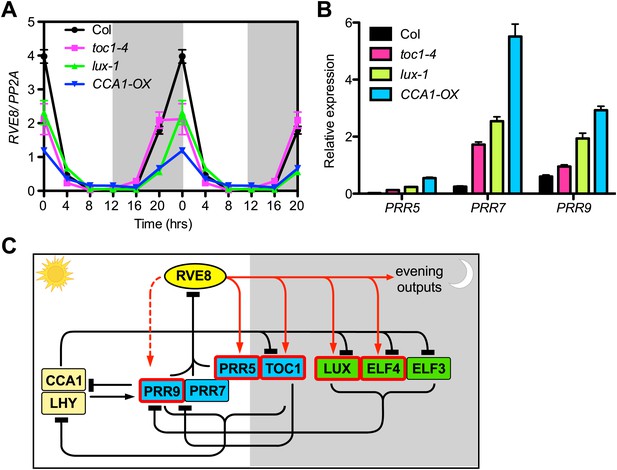
RVE8 expression is likely controlled by other clock genes through PRR5, 7 and 9.
(A) RVE8 expression in Col, toc1-4, lux-1 and CCA1-OX in LD. 7-day-old seedlings were collected at the times indicated and qRT-PCR was performed. Data are double-plotted to facilitate visualization. Values represent mean ± SEM. (B) Transcript levels of PRR5, PRR7 and PRR9 at ZT 0 (when RVE8 transcript levels normally peak) in wild-type (Col), toc1-4, lux-1, and CCA1-OX. Expression levels are normalized to PP2A. Data are represented as mean ± SEM from three technical replicates. (C) A proposed clock model integrating RVE8 as an activator of evening clock genes. The relative time of action of each component during diurnal cycles is shown from left to right. White box: day, grey box: night. REVEILLE/CCA1/LHY family proteins are shown in yellow; pseudo-response regulators are shown in blue; the evening complex components are shown in green. Clock components with one or more EE in their promoter regions are marked with red boxes. Red solid arrow: activation, red dashed arrow: activation only displayed in specific condition (red arrows are based on the current study), black perpendicular bars: repression, black arrow: activation. In this study, we demonstrated that RVE8 directly activates multiple evening-phased clock and output genes and that RVE8 is regulated by TOC1, LUX and CCA1, likely indirectly through their control of PRR5, 7 and 9 expression. For clarity, only transcriptional regulation is represented.
Tables
Enrichment of EE, G-box-like and ME-like motifs in CCGs regulated by RVE8 compared to their occurrence in all CCGs previously defined as either evening-phased or morning-phased (Hsu and Harmer, 2012)
| (A) Evening-phased genes (CT 8 to CT 14) | ||||||
| Motif | Sequence | CCGs (2709 genes) | RVE8-induced CCGs (278 genes) | p | ||
| Genes with the motif | Coverage (%) | Genes with the motif | Coverage (%) | |||
| Short EE | AAATATCT | 794 | 29.3 | 152 | 54.5 | <2.2 × 10−16*** |
| Long EE | AAAATATCT | 444 | 16.4 | 104 | 37.5 | 2.06 × 10−15*** |
| EE-like | AATATCT | 1360 | 50.2 | 190 | 68.2 | 7.39 × 10−09*** |
| (B) Morning-phased genes (CT 20 to CT 2) | ||||||
| Motif | Sequence | CCGs (1572 genes) | RVE8-repressed CCGs (328 genes) | p | ||
| Genes with the motif | Coverage (%) | Genes with the motif | Coverage (%) | |||
| G-box-like | BACGTRD | 1187 | 75.5 | 266 | 81.0 | 0.0317* |
| ME-like | CCACA | 1429 | 90.9 | 308 | 93.9 | 0.08297 |
-
To determine whether the over-represented motifs found in RVE8 targets (Supplementary file 2) are enriched when compared to the morning-phased and evening-phased CCG groups, the number of genes containing the motif in each phase group was compared to that in the up- or down-regulated RVE8 targets. Fisher's exact test was performed to determine if the ratios in both groups are significantly different (*p<0.05; **p<0.01; ***p<0.001).
Additional files
-
Supplementary file 1
RNA-seq analysis of RVE8-regulated genes.
(A) Pipeline and summary of RNA-seq data analysis. (B) Pseudo-normalized counts for visualization. Counts for each library were normalized (Robinson et al., 2010) according to the library size and represented as counts per million. (C) Genes identified as significantly induced by RVE8 by RNA-seq. Genes significantly differentially expressed (adjusted p<0.01) between RVE8:GR + DEX and RVE8:GR + mock or between RVE8:GR + DEX and rve8 + DEX, but not between rve8 + DEX and rve8 + mock, are defined as RVE8-induced or RVE8-repressed targets. (D) Genes identified as significantly repressed by RVE8 by RNA-seq. Genes significantly differentially expressed (adjusted p<0.01) between RVE8:GR + DEX and RVE8:GR + mock or between RVE8:GR + DEX and rve8 + DEX, but not between rve8 + DEX and rve8 + mock, are defined as RVE8-induced or RVE8-repressed targets. (E) Genes identified as significantly induced by DEX by RNA-seq. Genes significantly differentially expressed between rve8 + DEX and rve8 + mock are defined as DEX-induced or DEX-repressed genes (adjusted p<0.01). (F) Genes identified as significantly repressed by DEX by RNA-seq. Genes significantly differentially expressed between rve8 + DEX and rve8 + mock are defined as DEX-induced or DEX-repressed genes (adjusted p<0.01). (G) Clock genes identified as RVE8 targets in the RNA-seq experiment. Genes found to be differentially expressed in response to RVE8 induction (adjusted p<0.01). Evening-phased genes are highlighted in yellow. (H) Significantly overrepresented functional classifications among RVE8-induced genes. The enrichment of the functional terms was identified using BioMaps in Virtual Plant 1.2 (http://virtualplant.bio.nyu.edu/cgi-bin/vpweb/) (Katari et al., 2010). Functional classifications were provided by the Munich Information Center for Protein Sequences (MIPS) (Schoof et al., 2005). All genes classified as expressed in the RNA-seq experiment were used for the background. Fisher's exact test (with FDR correction) was performed and the cut-off value for statistical significance was set to 0.01.
- https://doi.org/10.7554/eLife.00473.014
-
Supplementary file 2
Promoter motifs overrepresented in RVE8-regulated CCGs (related to Table 1).
Promoters were defined as the 1500 bp region upstream of the translational start site and motifs were identified using the SCOPE motif finder (Carlson et al., 2007). Both strands were considered for calculation of significance. Background frequency was determined using all genes in the genome. (A) Up-regulated CCGs (376 genes). (B) Down-regulated CCGs (525 genes).
- https://doi.org/10.7554/eLife.00473.015
-
Supplementary file 3
Primers used in this study.
- https://doi.org/10.7554/eLife.00473.016





