The homologous recombination machinery modulates the formation of RNA–DNA hybrids and associated chromosome instability
Figures
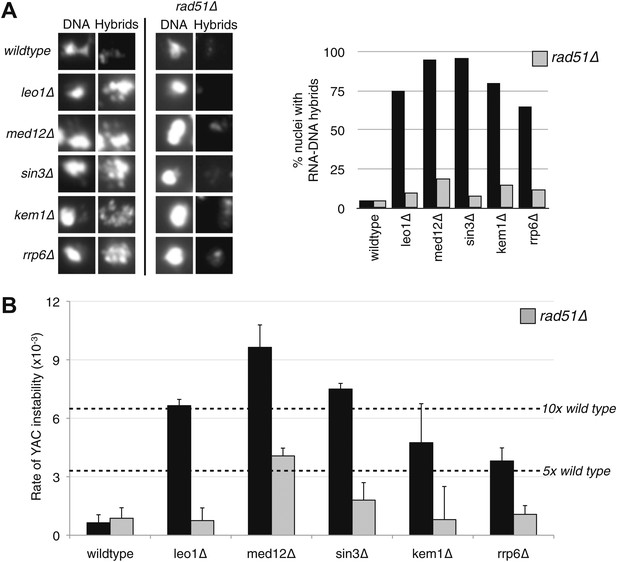
Deletion of RAD51 suppresses RNA–DNA hybrids and YAC instability.
(A) Left panel: Representative images of chromatin spreads stained with S9.6 antibody, showing reduced RNA–DNA hybrid staining in mutants with a deletion of RAD51 (rad51Δ). Right panel: The percent of total nuclei scored that stain positively for RNA–DNA hybrid in chromatin spreads is quantified. A total of 50–100 nuclei from two independent experiments were scored for each genotype. (B) Rate of yeast artificial chromosome (YAC) instability in mutants is also reduced when RAD51 is deleted. Error bars represent standard deviation calculated from at least four independent colonies.
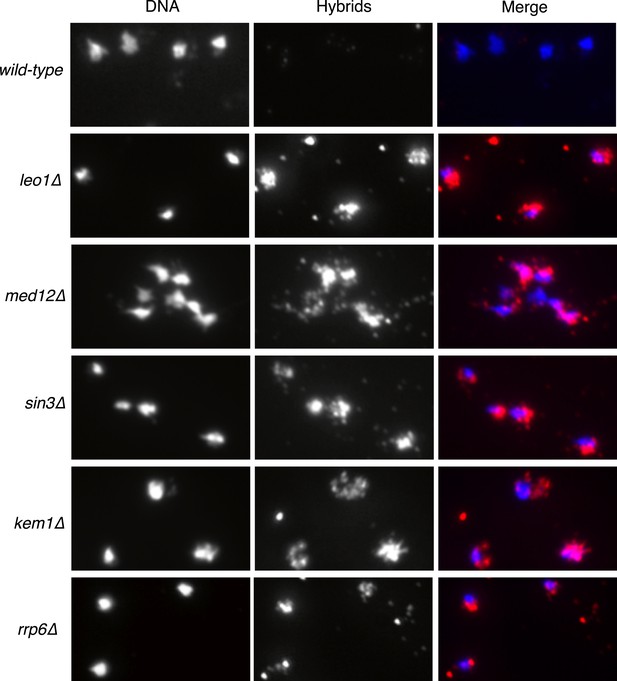
Larger panels of chromatin spreads showing multiple nuclei of single mutants stained with S9.6 antibody.
https://doi.org/10.7554/eLife.00505.004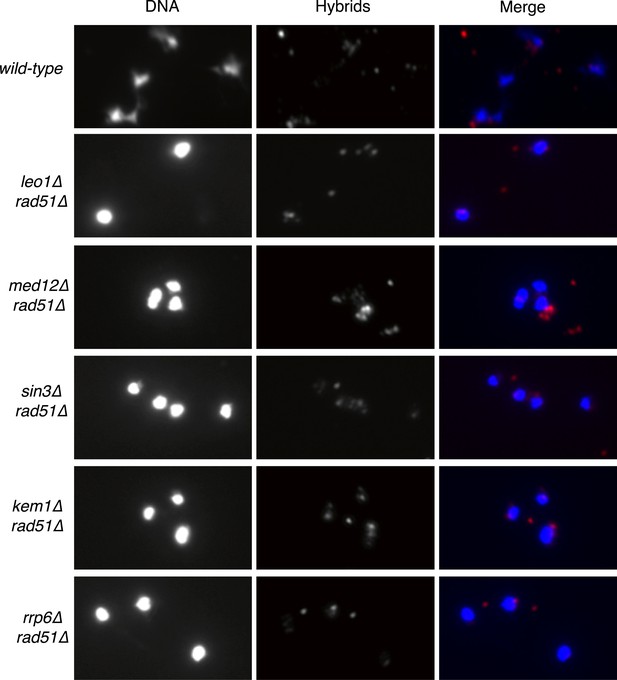
Larger panels of chromatin spreads showing multiple nuclei of double mutants stained with S9.6 antibody.
https://doi.org/10.7554/eLife.00505.005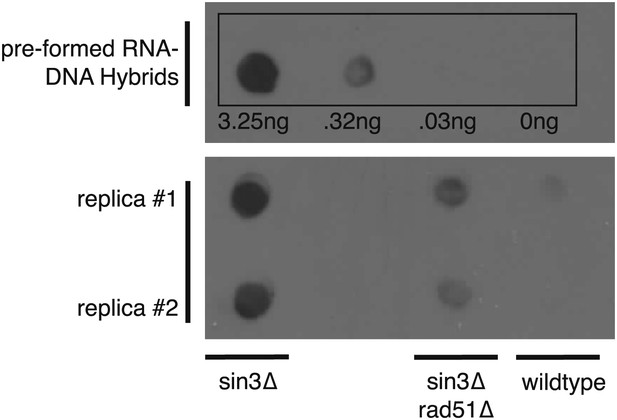
Dot blotting with S9.6 antibody.
Roughly 1 μg of DNA from indicated genotypes was spotted onto the membrane and stained with the S9.6 antibody. As a reference known amounts of pre-formed RNA–DNA hybrids were also spotted. Pre-formed RNA–DNA hybrids were made by performing a first strand synthesis reaction on total RNA. Amounts were quantified using Quant-iT Picogreen (Invitrogen, Carlsbad, CA).
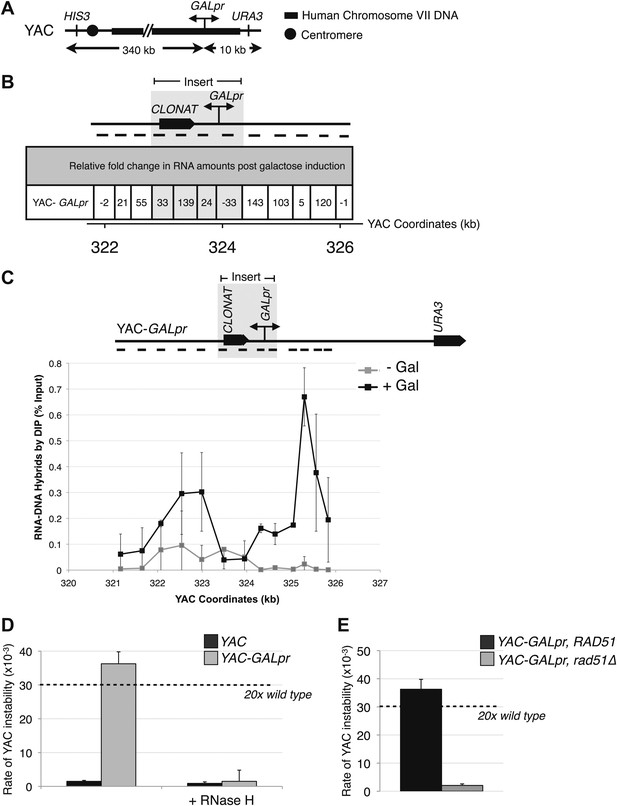
Hybrid-mediated YAC instability is induced in wild-type when high rates of transcription are induced on the YAC using the GAL1-10 promoter (GALpr).
(A) Schematic of the YAC-GALpr construct. Total yeast artificial chromosome (YAC) length is 350 kb, of which 324 kb come from human chromosome VII. The GALpr was integrated 10 kb from the telomere, on the arm with the URA3 marker. (B) Quantitative RT-PCR monitoring changes in RNA levels on the YAC 5 hr after induction with galactose. YAC RNA is normalized to actin RNA, and represented as fold change, as compared to RNA levels detected in uninduced cells. Above the table is a schematic representation of the YAC region from which RNA is measured, with the qRT-PCR fragments used in quantification indicated with black dashes. The region in gray represents the GAL1-10 promoter and selectable marker integrated in the YAC-GALpr strain. (C) DIP analysis to monitor RNA–DNA hybrid formation in the YAC-GALpr strain in the absence of galactose, and 2 hr after induction with galactose. Error bars represent standard deviation calculated from two independent DIP experiments. (D) Rates of YAC instability in strains with YAC (black bars) or YAC-GALpr (gray bars) 5 hr after addition of galactose to the media. Strains carried either an RNase H over-expressing plasmid or an empty control vector. (E) Induced YAC instability is suppressed when RAD51 is deleted. Error bars represent standard deviation calculated from at least three independent colonies.
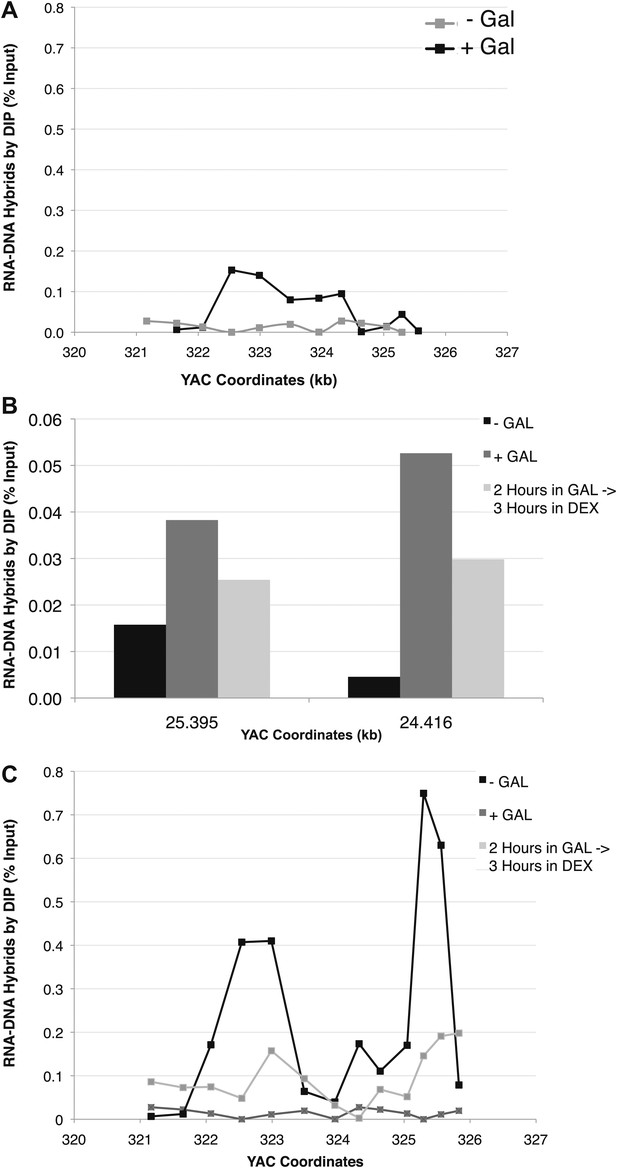
(A) DIP analysis of YAC strain prior to and 2 hr after addition of galactose to the media. (B) Monitoring of DIP signal in the YAC-GALpr strain at a distal region, showing low levels of hybrid signal upon induction with galactose as compared to. (C) DIP signals are reduced around the YAC-GALpr module upon return to repressive conditions.
https://doi.org/10.7554/eLife.00505.008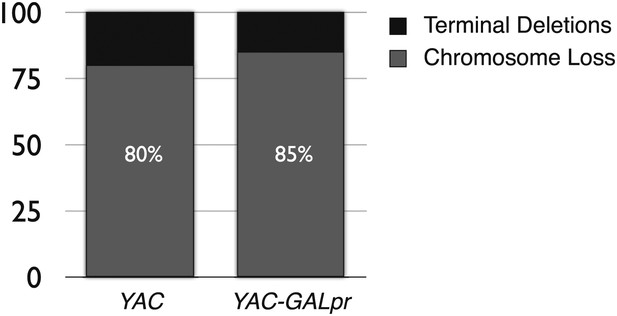
The percent of terminal deletions and chromosome loss events recovered after 5 hr of growth in galactose-containing media is comparable for YAC and YAC-GALpr strains.
https://doi.org/10.7554/eLife.00505.009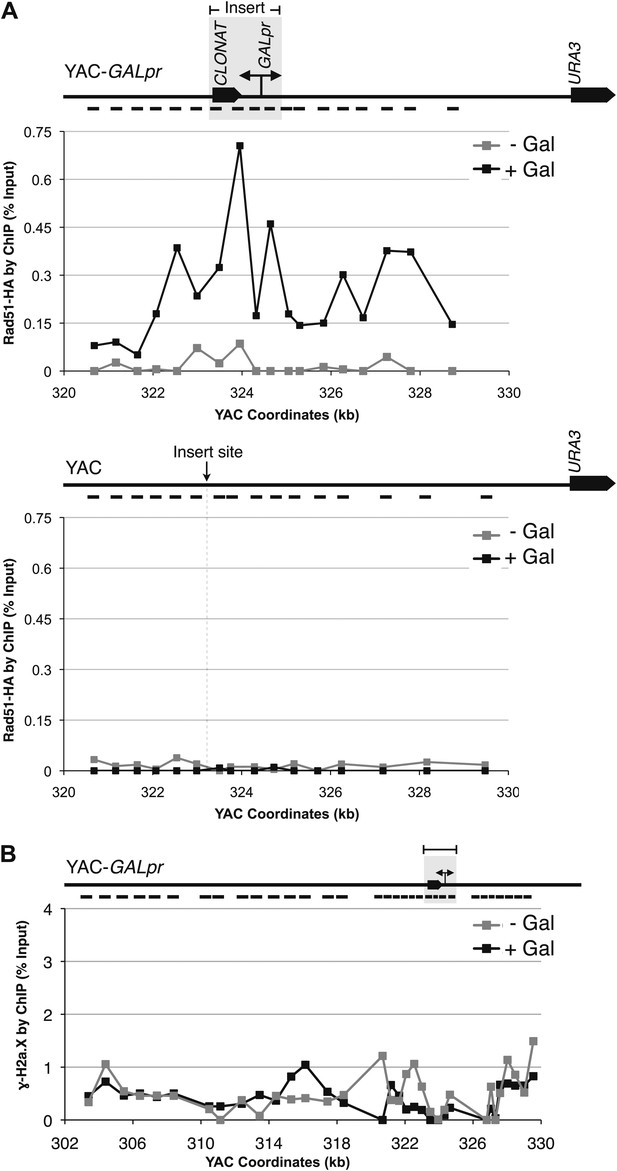
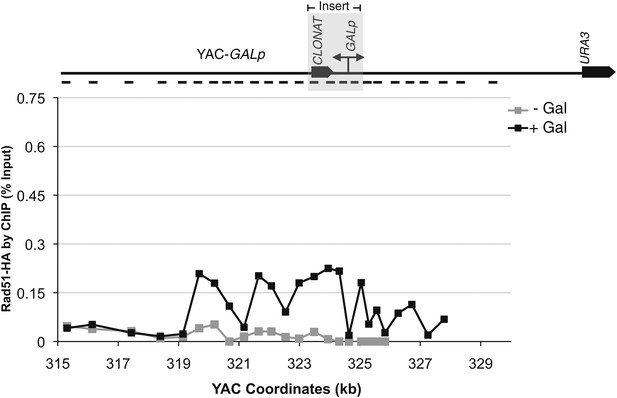
Rad51p binding is detectable around the YAC-GALpr module upon induction of transcription.
Cells growing exponentially in YEP-lactic acid were split, and galactose added to one half. The other half was collected immediately for the –Gal sample and fixed for chromatin immunoprecipitation (ChIP; see ‘Materials and methods’). After 120 min, the +Gal sample was similarly fixed for ChIP. Input DNA and DNA coimmunprecipitated with α-HA or -γ-H2a.X (IP) antibody were amplified using primer sets along the yeast artificial chromosome (YAC) as annotated with black dashes on the YAC-GALpr or YAC schematic above each graph. (A) ChIP of Rad51-HA in the YAC-GALpr strain shows an increased signal in Rad51-HA binding 2 hr after induction of transcription by addition of galactose to the media (top panel). No change in the RAD51-HA signal is observed in the YAC strain (bottom panel). (B) ChIP of γ-H2a.X in YAC-GALpr reveals no significant change in signal within 2 hr of galactose induction.
Rad51p binding is detectable around the YAC-GALpr module upon induction of transcription.
Chromatin immunoprecipitation (ChIP) of untagged Rad51 using a polyclonal antibody, showing an increased signal in Rad51 binding 2 hr after induction of transcription with galactose.
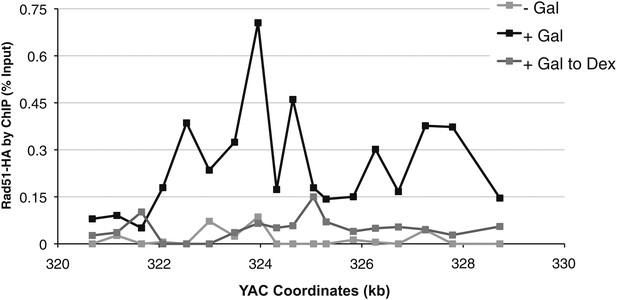
Rad51p binding is reduced around the YAC-GALpr module upon return to repressive conditions.
After 2 hr of growth in galactose-containing media, dextrose was then added to the media, and cells were allowed to grow, maintained in exponential phase, for 3 hr. Cells were then fixed and used for chromatin immunoprecipitation (ChIP) of Rad51-HA.
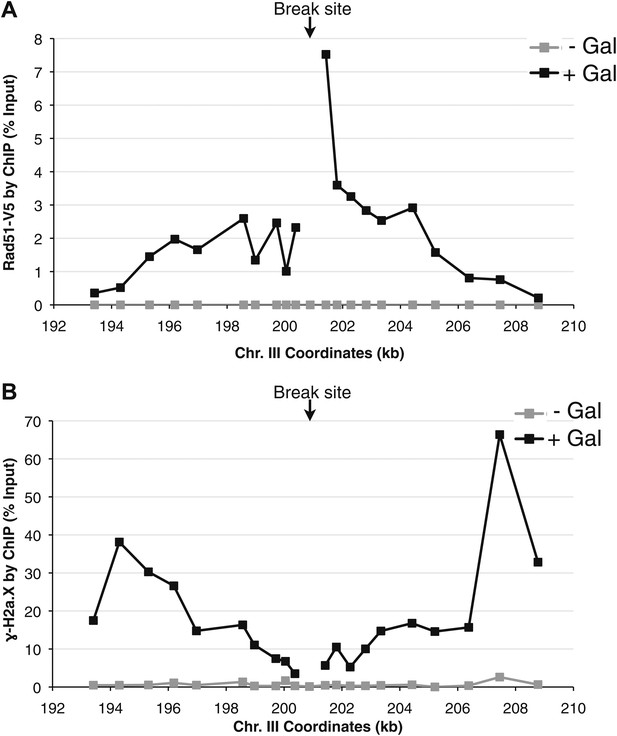
Rad51 and γ-H2a.X binding at an inducible break site on chromosome III.
(A) Chromatin immunoprecipitation (ChIP) of Rad51-V5 around a double-strand break induced by a site-specific HO endonuclease under control of a galactose inducible promoter. The graph shows levels of Rad51-V5 binding prior to, and 2 hr after galactose was added to the media. (B) ChIP of γ-H2a.X around the break site prior to, and 2 hr after galactose was added to the media.
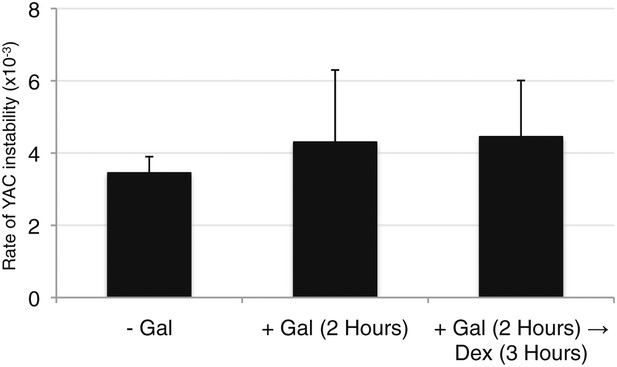
Wild-type levels of YAC instability are observed after 2 hr of transcription induction.
YAC-GALpr strains were grown for 2 hr in galactose-containing media, followed by addition of dextrose and growth for 3 more hours. Error bars represent standard deviation calculated from two independent colonies.
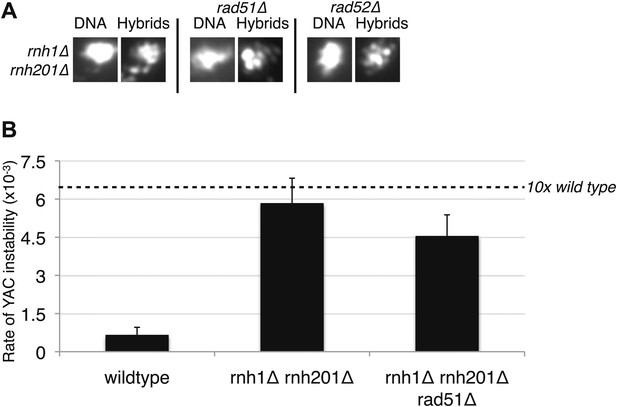
Deletions of RAD51 and RAD52 do not affect RNA–DNA hybrid formation in rnh1Δrnh201Δ.
(A) Representative images of chromatin spreads stained with S9.6 antibody. (B) Rate of yeast artificial chromosome (YAC) instability is similar in rnh1Δrnh201Δ and strains lacking RAD51 (rad51Δ) or RAD52 (rad52Δ). Error bars represent standard deviation calculated from at least six independent colonies.
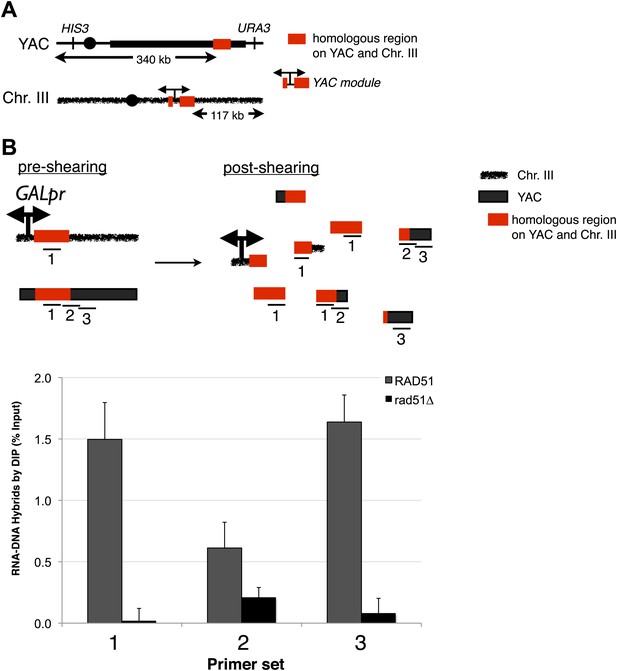
Transcription of YAC sequences from chromosome III causes RNA–DNA hybrid formation in trans on the YAC.
(A) Schematic representation of the trans assay is depicted. The GALpr, selectable marker (CLONAT), and 1.1 kb of yeast artificial chromosome (YAC) DNA was integrated on chromosome III. (B) Schematic representation of where the primer sets used to monitor hybrid formation in trans are depicted. Hybrid formation is monitored by DIP 2 hr after induction with galactose in RAD51 and rad51Δ strains. Error bars represent standard deviation from two independent experiments.
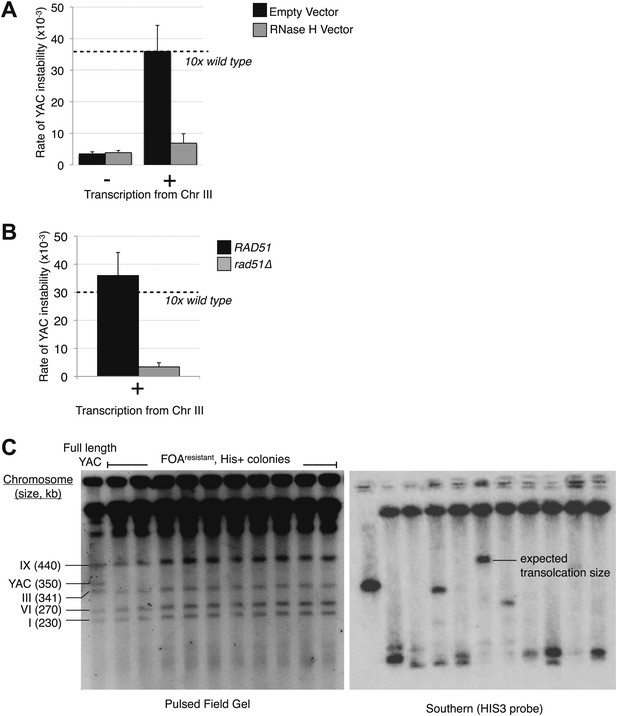
Transcription of YAC sequences in trans causes hybrid-mediated YAC instability.
(A) Rates of yeast artificial chromosome (YAC) instability in strains carrying an empty control vector (black bars) or RNase H over-expressing vector (gray bars) showing an increased rate of instability upon induction of transcription that is reduced when RNase H is over-expressed. Error bars represent standard deviation calculated from at least three independent colonies. (B) Rate of YAC instability is suppressed when RAD51 is knocked out. (C) Pulse-field gel and Southern analysis with HIS3 probe of FOAresistant, His+ colonies, showing that 9/10 colonies analyzed have YACs rearranged to a smaller size.
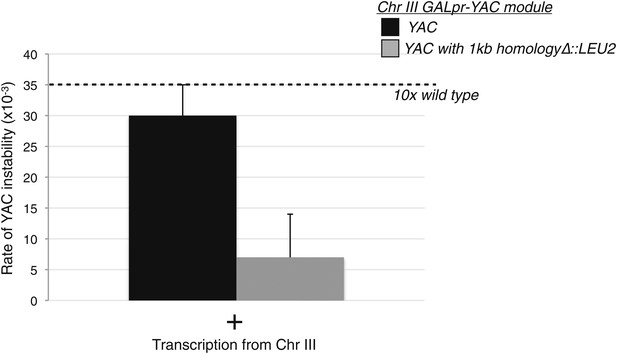
Levels of YAC instability in the trans assay with and without a region of homology on the YAC.
The 1 kb region of the yeast artificial chromosome (YAC) inserted on chromosome III (YAC-GALpr module) was replaced on the YAC with a LEU2 cassette. Wild-type levels of YAC instability are observed in that strain after 2 hr of induction with galactose.
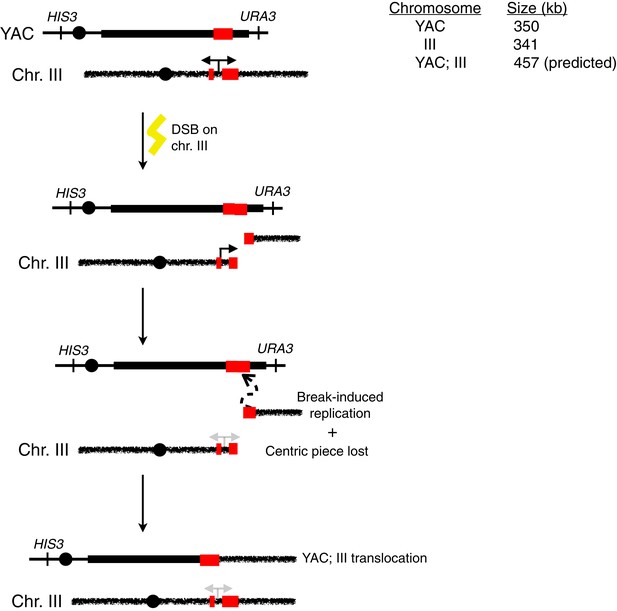
Schematic representation of an alternative for how HIS+ URA− colonies may arise in the trans assay.
Upon induction of transcription, breaks may occur in cis on chromosome III. Repair via break-induced replication using homologous yeast artificial chromosome (YAC) sequences as a substrate can lead to a chromosome III;YAC translocation.
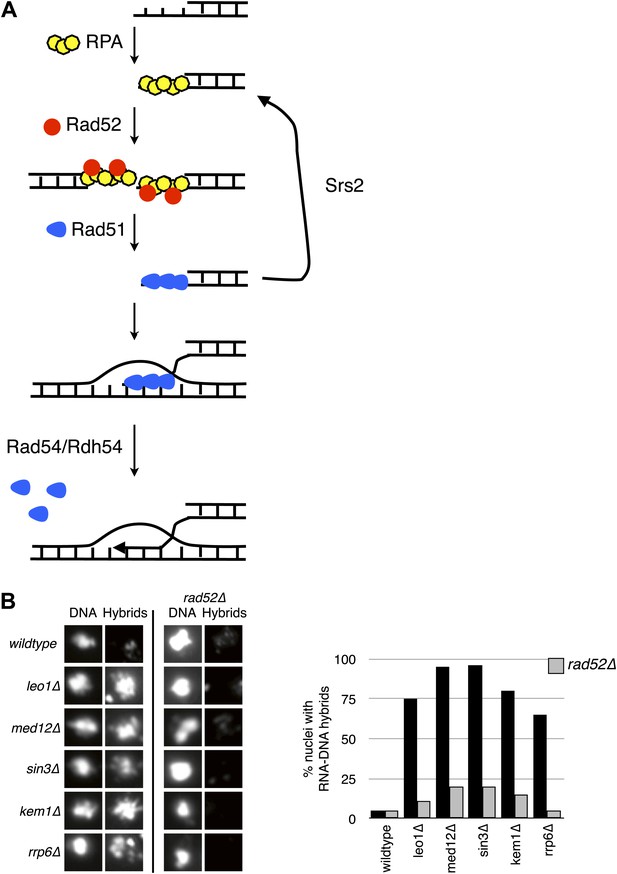
Deletion of RAD52 suppresses RNA–DNA hybrids.
(A) Schematic showing the major proteins canonically involved in regulating Rad51p binding in DNA repair. Following resection, replication protein A (RPA) polymerizes onto ssDNA. Rad52 then interacts with RPA and catalyzes its exchange for Rad51p. The Rad51–ssDNA filament promotes the pairing and strand exchange reaction with a homologous region in duplex DNA. Srs2p, Rad54p, and Rdh54p all regulate the Rad51 filament by dismantling Rad51 from ssDNA and dsDNA, respectively. (B) Representative images of chromatin spreads stained with S9.6 antibody and quantification of nuclei, showing reduced RNA–DNA hybrid staining in mutants with RAD52 knocked out. A total of 50–100 nuclei from two independent experiments were scored for each genotype.
Larger panels of chromatin spreads showing multiple nuclei of rad52Δ mutants stained with S9.6 antibody.
https://doi.org/10.7554/eLife.00505.021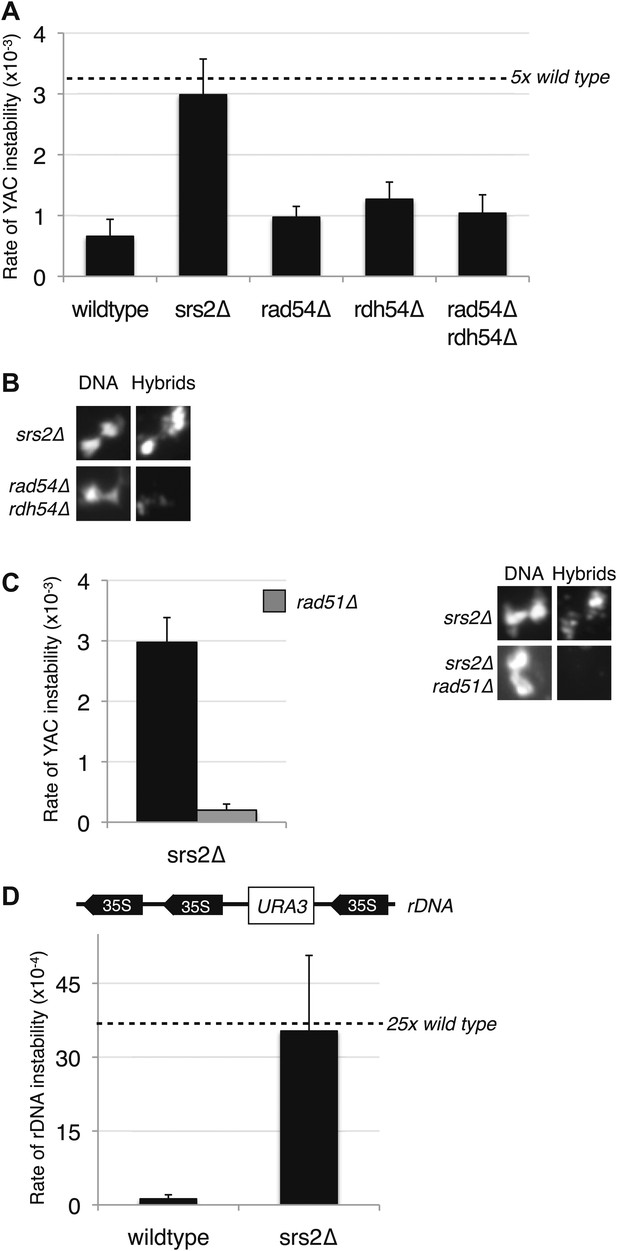
Deletion of SRS2, but not RAD54 and RDH54 increases genome instability and hybrid formation.
(A) Rate of yeast artificial chromosome (YAC) instability is increased in srs2Δ but not in rad54Δ and rdh54Δ single and double mutants. Error bars represent standard deviation calculated from at least six independent colonies. (B) Representative images of chromatin spreads stained with S9.6 antibody, showing increased RNA–DNA hybrid staining in srs2Δ. (C) Left panel: Rate of YAC instability in srs2Δ is suppressed when RAD51 is knocked out. Right panel: Hybrid staining is also reduced in the srs2Δ rad51Δ double mutant. (D) srs2Δ mutants with URA3 integrated at the rDNA were assayed for loss of the URA3 marker, showing increased instability. Error bars represent standard deviation calculated from at least six independent colonies.
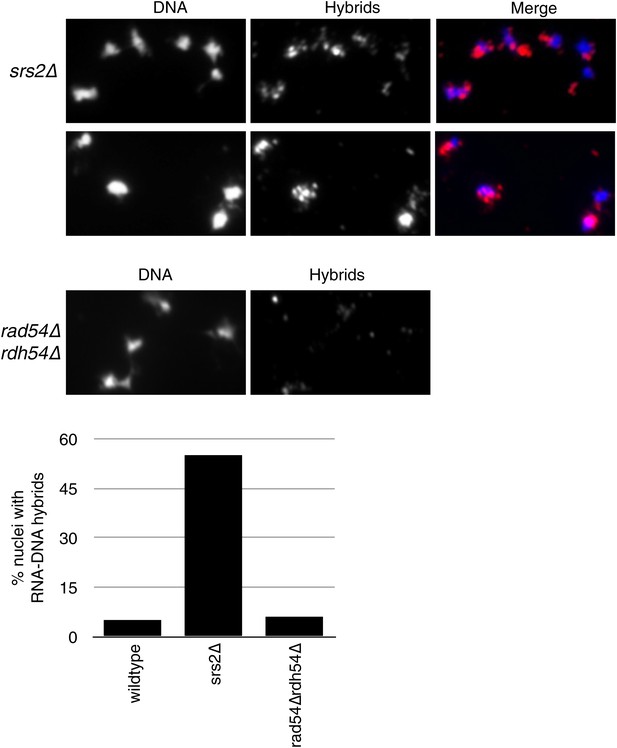
Larger panels of chromatin spreads showing multiple nuclei of srs2Δ and rdh54Δrad54Δ mutants stained with S9.6 antibody.
Quantification of number of nuclei that stained positively for RNA–DNA hybrids in srs2Δ and rdh54Δrad54Δ. Total number of nuclei scored is 50–100 per genotype, from two independent experiments.
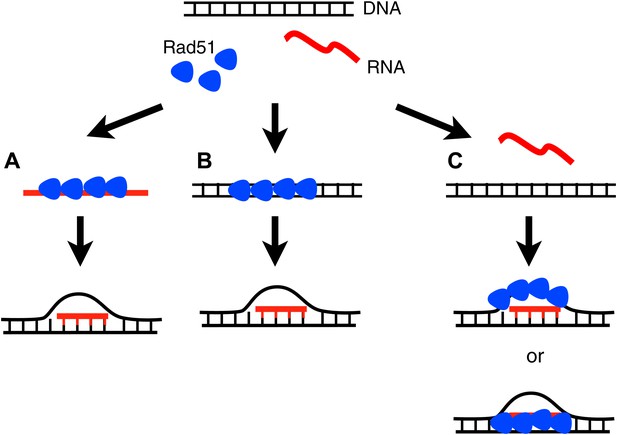
Three models for how Rad51p may mediate RNA–DNA hybrid formation.
(A) In the forward reaction, Rad51p polymerizes onto RNA, and mediates strand exchange with homologous DNA, forming an RNA–DNA hybrid. (B) In the inverse reaction, Rad51p forms a filament on dsDNA and promotes strand exchange with homologous RNA. (C) A third alternative is that Rad51 forms a filament on the extruded ssDNA, stabilizing an open D-loop that allows RNA to bind to homologous sequences.
Additional files
-
Supplementary file 1
(A) Description of strains used in this study. (B) Chromatin immunoprecipitation (ChIP)/DNA immunoprecipitation (DIP) primers and integration primers for integration of the GALpr in LW6811.
- https://doi.org/10.7554/eLife.00505.025





