A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics
Figures
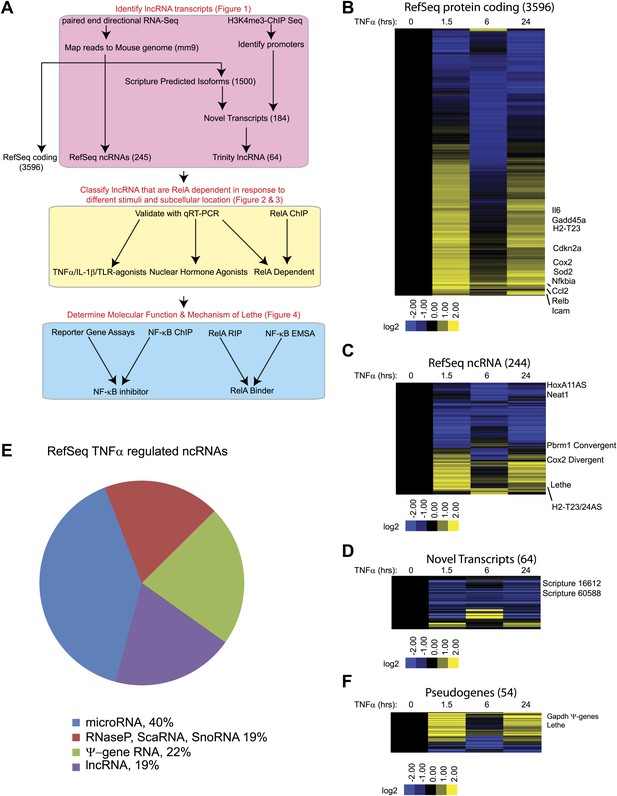
TNFα regulates the transcription of many coding and noncoding genes.
(A) Workflow for strategy for discovery of NF-κB regulated lncRNAs. (B) 3596 RefSeq protein coding genes are regulated by TNFα. Values are normalized to the 0 hr time point. (C) 244 RefSeq ncRNAs are regulated by TNFα. (D) 64 de novo lncRNAs are regulated by TNFα. (E) The fraction of all RefSeq ncRNAs for each class of transcript. (F) 54 pseudogene lncRNAs are regulated by TNFα.
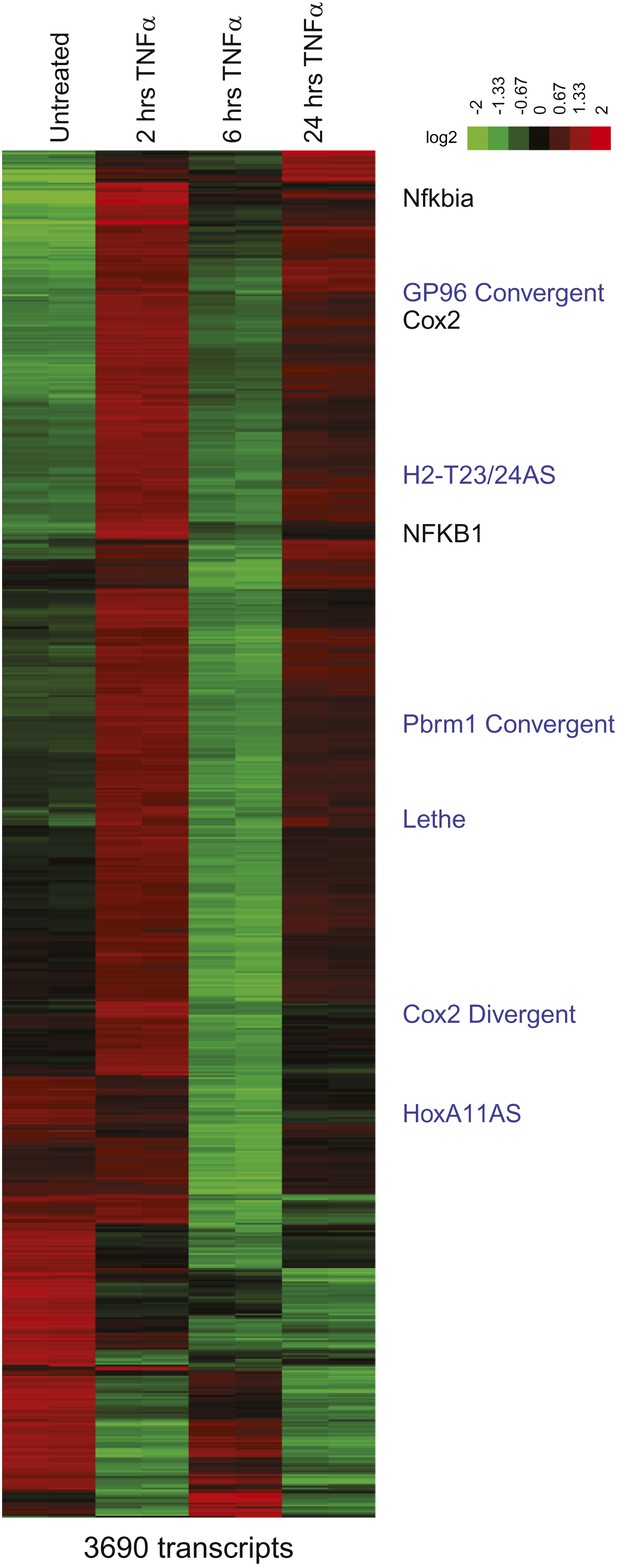
Heatmap of RefSeq genes.
Mean centered heatmap of RefSeq protein coding genes and RefSeq lncRNAs.
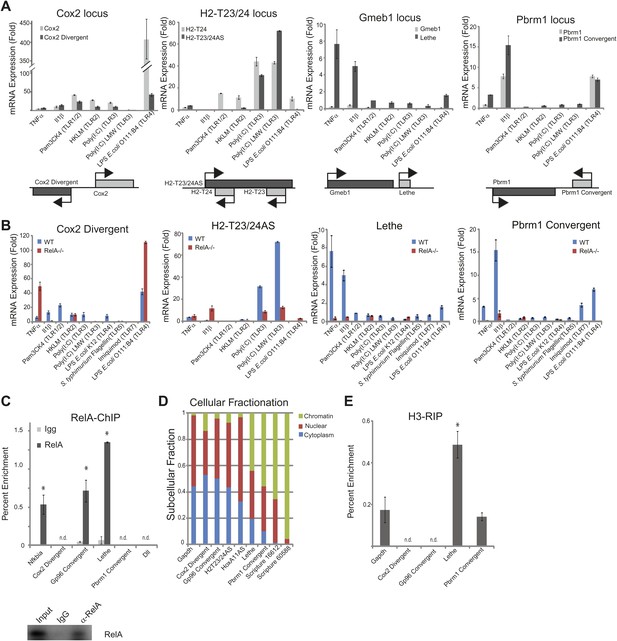
LncRNAs distinguish between different stimuli and are regulated by NF-κB.
(A) Validation of lncRNAs expression alongside the closest protein coding gene under a variety of different stimuli by qRT-PCR. Genomic organization is shown below. MEFs were treated with 20 ng/ml TNFα for 0 and 6 hr. Quantitative Taqman real time RT-PCR of the indicated RNAs is normalized to Actin levels (mean ± SD). (B) LncRNAs are regulated by RelA. qRT-PCR in WT and RelA−/− littermate cells under a variety of different stimuli. MEFs were treated with 20 ng/ml TNFα for 0 and 6 hr. Quantitative Taqman real time RT-PCR of the indicated RNAs is normalized to Actin levels (mean ± SD). (C) Endogenous RelA is recruited to the promoters of lncRNAs. MEFs were treated with 20 ng/ml TNFα for 0 and 15 min. ChIP with α-RelA antibodies was performed and RelA percent enrichment relative to input is shown (mean ± SD, Nkfbia, p<0.0518; Gp96 Convergent, p<0.007; Lethe, p<0.002). (D) LncRNAs are found throughout the cell. Cellular fractionation was performed and fraction found in the chromatin, nucleus and cytoplasm is shown. MEFs were treated with 20 ng/ml TNFα for 6 hr. Quantitative Taqman real time RT-PCR of the indicated RNAs is shown (mean ± SD is shown). (E) LncRNAs are found on the chromatin. MEFs were treated with 20 ng/ml TNFα for 6 hr. RNA-IP with α-H3 antibodies was performed. RNA was isolated and Quantitative Taqman real time RT-PCR of the indicated RNAs is shown (mean ± SD, Lethe p<0.004).
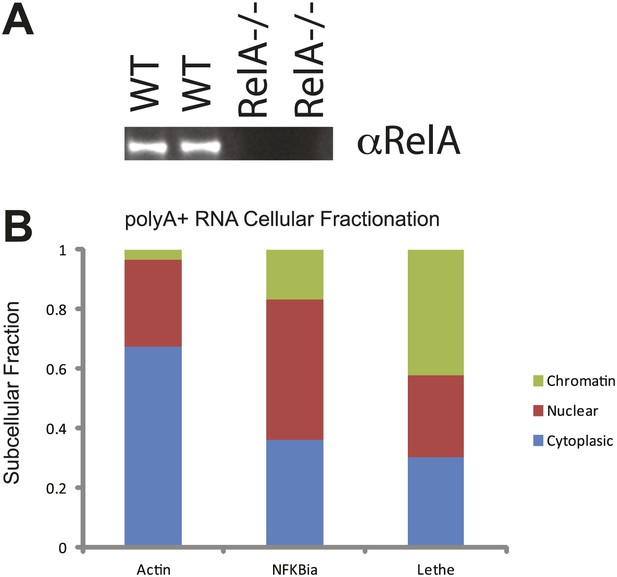
(A) Western analysis of RelA protein levels. (B) PolyA+ Lethe is found on the chromatin.
Immunoblot of wildtype and RelA−/− MEFs. Cellular fractionation was performed, total RNA was purified and polyA+ selection was performed. The fraction polyA+ RNA found in the chromatin, nucleus and cytoplasm is shown. MEFs were treated with 20 ng/ml TNFα for 6 hr. Quantitative Taqman real time RT-PCR of the indicated RNAs is shown (mean ± SD is shown).
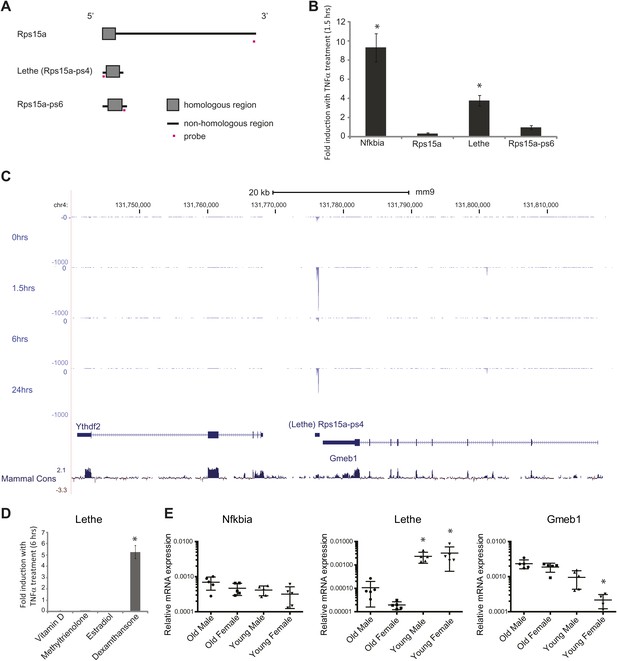
Lethe is a pseudogene lncRNAs that is regulated by NF-κB, Glucocorticoid Receptor and in aging.
(A) Gene structure, homology and Taqman probe design of Rps15a and pseudogene family members. (B) Lethe is induced by TNFα, but other family members are not. MEFs were treated with 20 ng/ml TNFα for 0 and 1.5 hr. Quantitative Taqman real time RT-PCR of the indicated RNAs is shown normalized to Actin levels (mean ± SD, p<0.012). (C) Genomic organization of Lethe with RNA-Seq data at time 0, 1.5, 6 and 24 hr post TNFα treatment. Lethe is located on mouse chromosome 4 between Gmeb1 and Ythd2. Gmeb1 and Ythd2 are not induced by TNFα stimulation. (D) Lethe is induced by dexamethasone treatment, but not other nuclear hormone receptor agonists. MEFs were treated with either 10 nM vitamin D, 100 nM methyltrienolone, 100 nM estradiol, or 1 μM dexamethasone for 0 and 6 hr. Quantitative Taqman real time RT-PCR of the indicated RNAs is shown normalized to Actin levels (mean ± SD, p<0.003). (E) Lethe is down-regulated in aged mice. Lethe is expressed in young spleen from male and female mice. Five mice were used for each sex and time point. Quantitative Taqman real time RT-PCR of the indicated RNAs is shown normalized to Actin levels (mean ± SD, Lethe p<0.001, Gmeb1 p<0.003). ANOVA analysis was performed to determine significance.

Alignment of Lethe with Rps15a-ps6.
ClustalW2 alignment was performed on Lethe (Rps15a-ps4) and Rps15a-ps6.
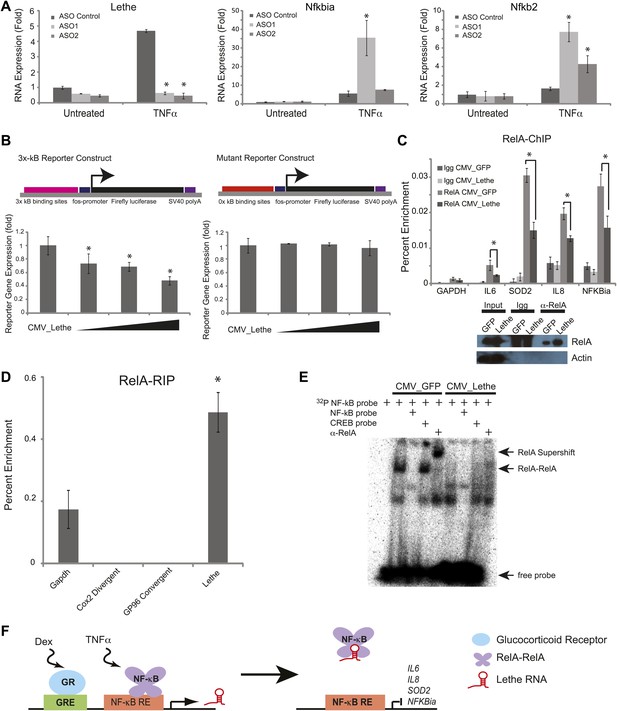
Lethe Binds to RelA and inhibits RelA occupancy of DNA.
(A) Increased expression of NF-κB regulated genes in Lethe knockdown cells. Quantitative Taqman real time RT-PCR of the indicated RNAs is shown normalized to Actin levels (mean ± SD, p<0.05 is shown) (B) Lethe inhibits TNFα induced reporter gene expression. RLU of 3x-κB reporter activity and mutant reporter activity (mean ± SD, p<0.05 is shown) in CMV_Lethe transfected 293T cells. Reporter constructs are diagrammed above. (C) Endogenous RelA recruitment to the promoters of target genes is reduced in the presence of Lethe. 293T expressing CMV_GFP or CMV_Lethe were treated with 20 ng/ml TNFα for 15 min. ChIP with α-RelA antibodies was performed and RelA percent enrichment relative to input is shown (mean ± SD; Il6, p<0.033; Sod2, p<0.001; Il8, p<0.003; Nfkbia, p<0.015). (D) Lethe binds to RelA. MEFs were treated with 20 ng/ml TNFα for 6 hr. RNA-IP with α-RelA antibodies was performed. RNA was isolated and Quantitative Taqman real time RT-PCR of the indicated RNAs is shown (mean ± SD, p<0.020). (E) Lethe expression blocks DNA binding of the RelA homodimer to its target. NF-κB EMSA of GFP or Lethe transfected 293T nuclear extracts treated with 20 ng/ml TNFα for 15 min. Extracts were pretreated with unlabeled NF-κB (specific) or CREB (nonspecific competitor), or α-RelA antibodies for 15 min prior to incubation with probe. (F) Model for Lethe regulation of gene expression. Upon addition of TNFα or dexamthasone, Lethe is transcribed. Lethe can then bind to RelA–RelA homodimers and block binding to other NF-κB response elements, inhibiting NF-κB.
Tables
Classification of TNF regulated lncRNAs
| TNFα | IL-1β | TLR1 | TLR2 | TLR3 | TLR4 | TLR5 | TLR7 | RelA-dep | H3-RIP | |
|---|---|---|---|---|---|---|---|---|---|---|
| Cox2 Divergent | + | + | + | + | + | + | n.d. | n.d. | − | n.d. |
| Gp96 Convergent | + | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | + | n.d. |
| H2-T23/24AS | + | n.d. | n.d. | n.d. | + | n.d. | n.d. | n.d. | − | − |
| HoxA11AS | − | − | − | − | + | − | − | − | − | − |
| Lethe | + | + | − | − | − | − | − | − | + | + |
| Pbrm1 Convergent | + | + | − | − | − | − | − | + | −/+ | + |
| Scripture 16,612 | − | + | + | + | − | − | − | − | n.d. | n.t. |
| Scripture 60,588 | − | − | − | − | − | − | − | − | n.d. | n.t. |
-
These results demonstrate that lncRNAs are regulated by diverse and specific stimuli. Additionally, lncRNAs are directly regulated by NF-κB. Finallyp, the subcellular distribution of TNFα−induced lncRNAs varies by transcript. n.d., not detectable. n.t., not tested.
Additional files
-
Supplementary file 1
(A) Sequencing depth for all conditions. (B) List of de novo transcripts: Trinity generated transcripts with the genomics coordinates, the number of mapped reads, RPKMs, CPC score and if there is an H3K4me3 peak from ChIP-Seq experiments. (C) List of RefSeq coding genes: RefSeq genes with the number of mapped reads, RPKMs and fraction nucleotide coverage for each. (D) List of RefSeq ncRNA: RefSeq ncRNA with the number of mapped reads, RPKMs and nucleotide coverage for each. (E) List of pseudogenes: pseudogene ncRNA with the number of mapped reads, RPKMs and nucleotide coverage for each. (F) List of significantly expressed transcripts from RefSeq: RefSeq transcripts with the log2RPKM values. (G) List of all primers used: primer sequences used for ChIP, Taqman assays, antisense oligos, and primers for cloning.
- https://doi.org/10.7554/eLife.00762.011





