miR-146a promotes the initiation and progression of melanoma by activating Notch signaling
Figures
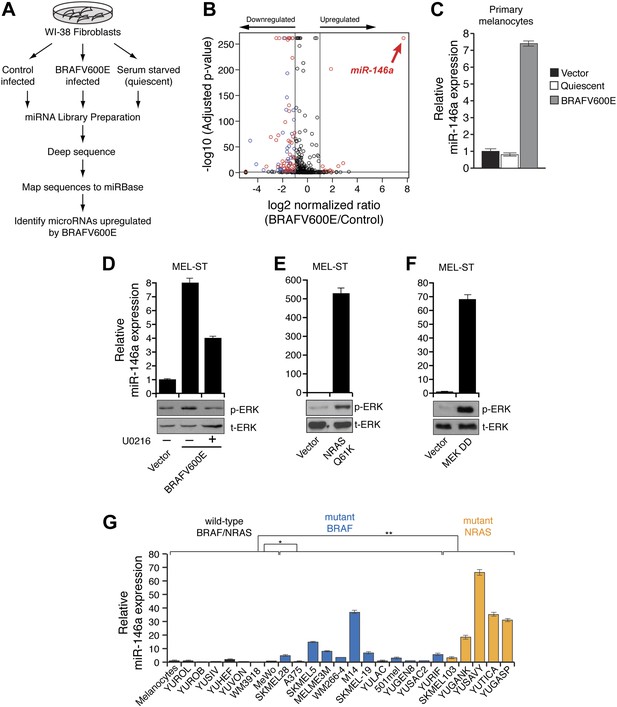
BRAFV600E and NRASQ61K upregulate miR-146a expression.
(A) Schematic summary of study design. (B) Volcano plot showing miR-146a (red arrow) as the most upregulated miRNA. (C) qRT-PCR analysis of miR-146a expression in primary melanocytes transduced with BRAFV600E (gray) or Vector control (black) retrovirus particles. Quiescent cells were used as a control (light gray). (D) qRT-PCR analysis (top) measuring miR-146a expression in MEL-ST cells expressing BRAFV600E in the presence (+) or absence (−) of the MEK inhibitor U0216 (10 µM) relative to Vector control. Immunoblots (bottom) of phosphorylated (p-) ERK (upper) and total (t-) ERK (lower) show that BRAFV600E activates MEK-dependent phosphorylation of ERK. (E) qRT-PCR analysis (top) measuring miR-146a expression in MEL-ST cells expressing NRASQ61K relative to Vector control. Immunoblots (bottom) of phosphorylated (p-) ERK (upper) and total (t-) ERK (lower) show that NRASQ61K activates phosphorylation of ERK. (F) qRT-PCR analysis (top) measuring miR-146a expression in MEL-ST cells expressing constitutively active MEK (DD), relative to the Vector control. Immunoblots (bottom) of p-ERK (upper) and t-ERK (lower) show that MEK (DD) activates phosphorylation of ERK. (G) qRT-PCR analysis monitoring miR-146a expression in melanocyte and melanoma cell lines. BRAF (blue) and NRAS (orange) mutant cell lines are indicated.
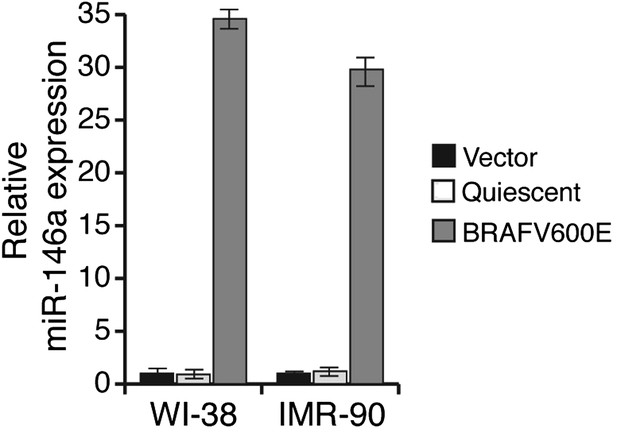
Ectopic expression of BRAFV600E in WI-38 and IMR-90 cells stimulates miR-146a expression.
qRT-PCR analysis of miR-146a expression in WI-38 or IMR-90 cells transduced with BRAFV600E (gray) or Vector control (black) retrovirus particles. Quiescent cells were used as a control (light gray).
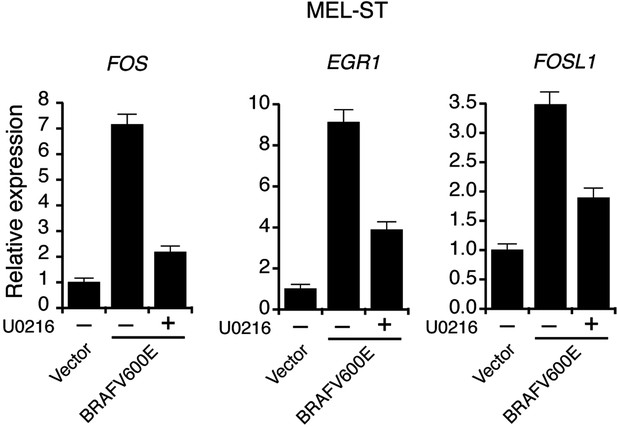
Activation of MAP Kinase pathway target genes upon ectopic expression of BRAFV600E in MEL-ST cells.
qRT-PCR analysis of MEK-ERK transcriptional targets FOS, EGR1 and FOSL1 under indicated conditions in MEL-ST cells expressing either an empty vector or BRAFV600E.
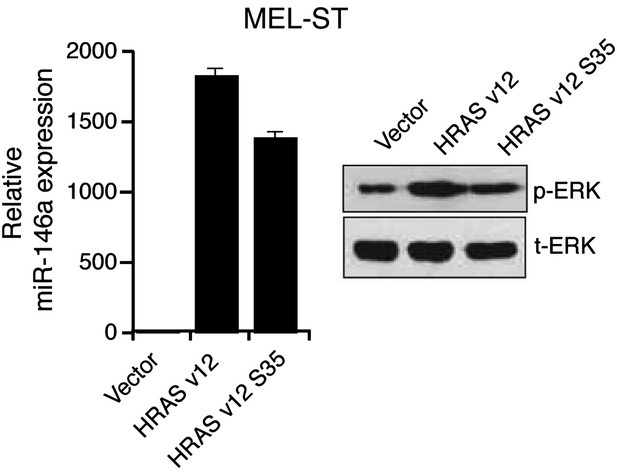
Ectopic expression of HRAS v12 or HRAS v12 S35 in MEL-ST cells stimulates miR-146 expression.
qRT-PCR analysis of miR-146a (left) and immunoblot analysis (right) of phosphorylated (p-) ERK, total (t-) ERK in MEL-ST cells transduced with empty vector, HRAS v12, or HRAS v12s35.
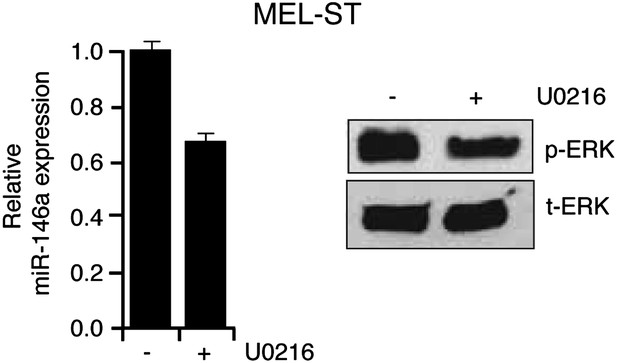
Inhibition of MAP kinase signaling by MEK inhibitor blocks HRAS v12-mediated miR-146a upregulation.
qRT-PCR analysis (left) to monitor miR-146a expression and immunoblot analysis (right) of phosphorylated (p-) ERK and total (t-) ERK in MEL-ST cells transduced with HRAS v12 in the presence (+) or absence (−) of the MEK inhibitor U0216.
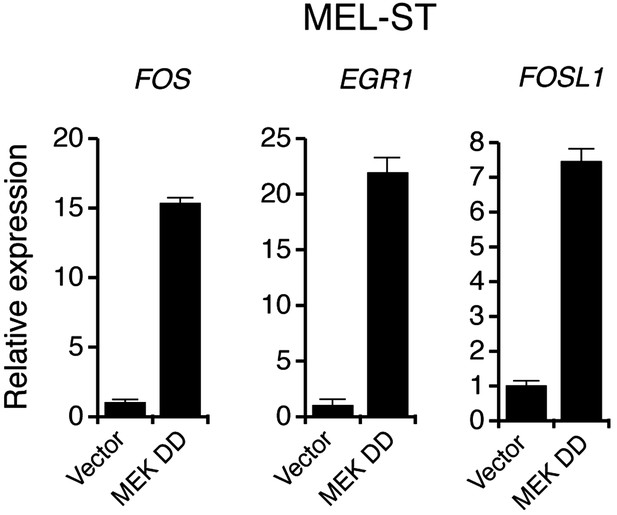
Ectopic expression of MEK DD stimulates the transcription of MAP kinase target genes.
qRT-PCR analysis of MEK-ERK transcriptional targets FOS, EGR1 and FOSL1 in MEL-ST cells expressing either a empty vector or constitutively active MEK (MEK DD).
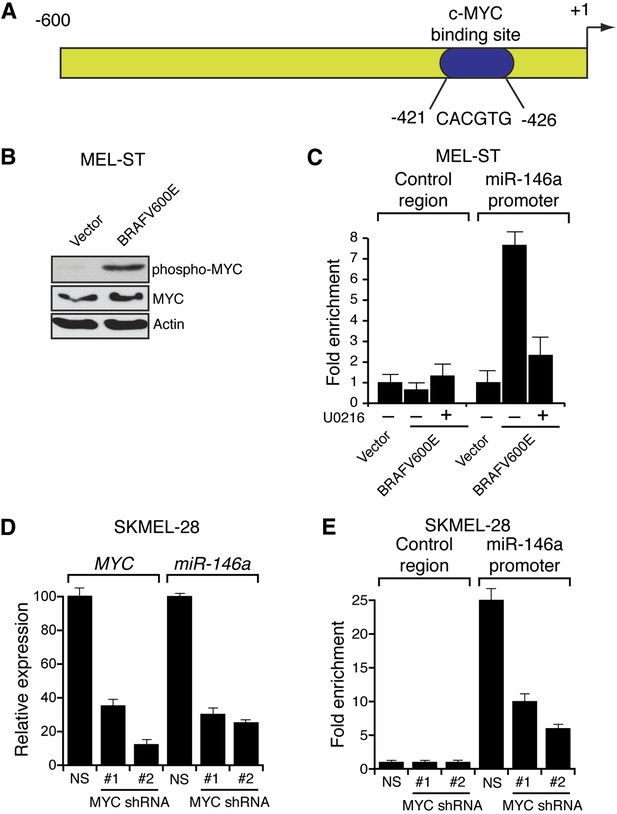
BRAFV600E upregulates miR-146a through MYC oncogene.
(A) Schematic representation of MYC promoter and the miR-146a binding site that has been identified by PROMO analysis software. ‘+1’ indicates the transcription start site. (B) Immunoblot analysis of p-MYC and total MYC in whole cell lysates of MEL-ST cells transduced with empty vector or BRAFV600E. Actin was used as a loading control. (C) Chromatin immunoprecipitation (ChIP) assay measuring MYC binding to the miR-146a promoter in MEL-ST cells stably expressing BRAFV600E in the presence (+) or absence (−) of the MEK inhibitor U0216 relative to cells transduced with the empty vector. A non-specific control region served as a negative control for MYC recruitment. (D) qRT-PCR analysis of MYC mRNA (left) and miR-146a (right) expression in SKMEL-28 cells infected with MYC shRNAs. (E) qPCR analysis of MYC ChIP of miR-146a promoter and negative control in SKMEL-28 cells infected with MYC shRNAs.
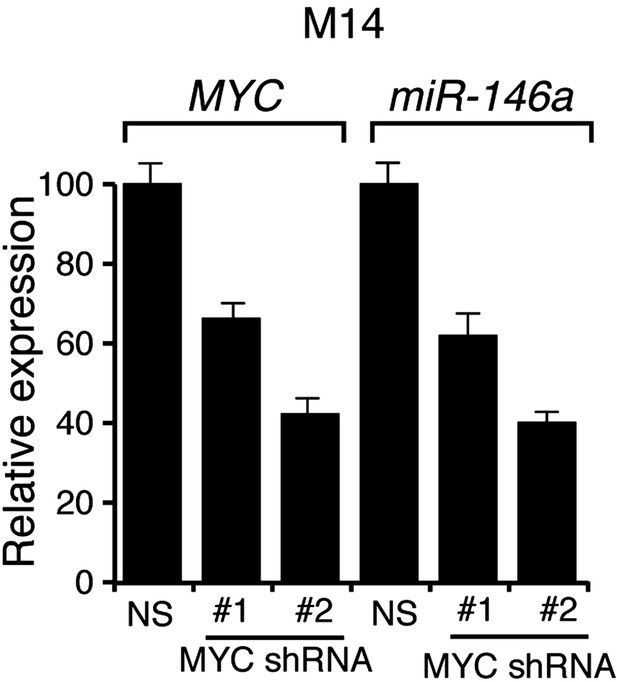
Analysis of MYC and miR-146a expression in M14 cells expressing shRNAs against MYC.
qRT-PCR analysis of MYC and miR-146a expression in M14 cells transduced with MYC shRNA expression vectors relative to cells transduced with a non-specific (NS) shRNA vector.
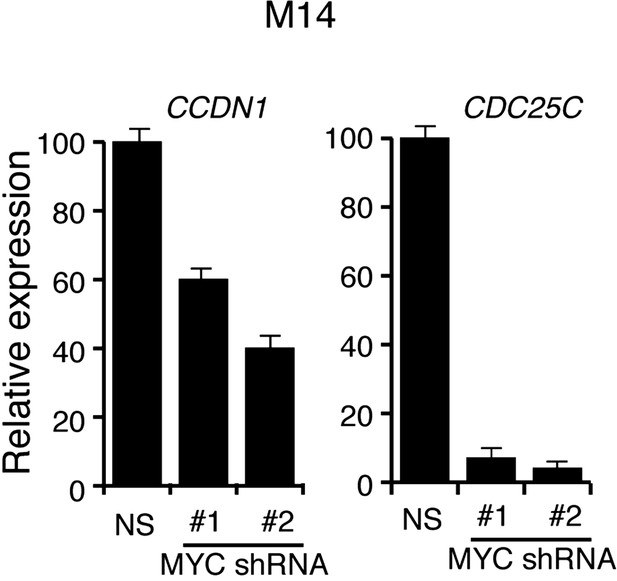
shRNA-mediated downrgulation of MYC in M14 cells inhibits the expression of MYC transcriptional target genes.
qRT-PCR analysis of MYC targets CCDN1 and CDC25C in M14 cells transduced with MYC shRNA expression vectors relative to cells transduced with a non-specific (NS) shRNA vector.
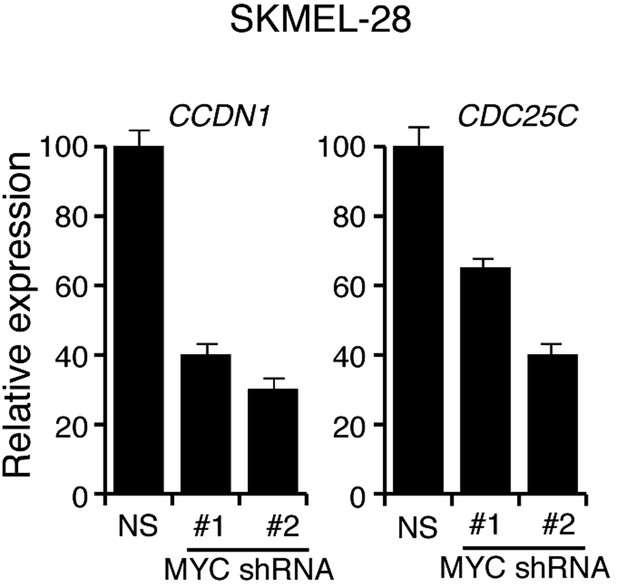
shRNA-mediated downrgulation of MYC in SKMEL-28 cells inhibits the expression of MYC transcriptional target genes.
qRT-PCR analysis of MYC targets CCDN1 and CDC25C in SKMEL-28 cells transduced with MYC shRNA expression vectors relative to cells transduced with a non-specific (NS) shRNA vector.
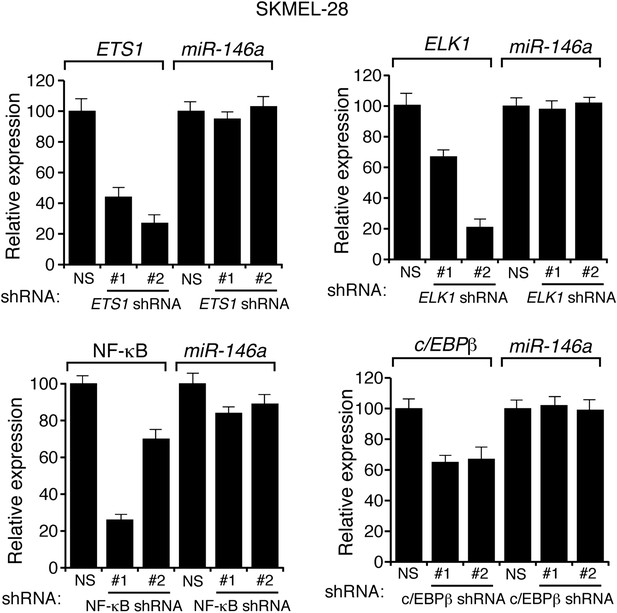
Transcriptional regulation of miR-146a.
qRT-PCR analysis of ETS1, ELK1, NF-κB, c/EBPβ and miR-146a in SKMEL-28 expressing indicated shRNA.
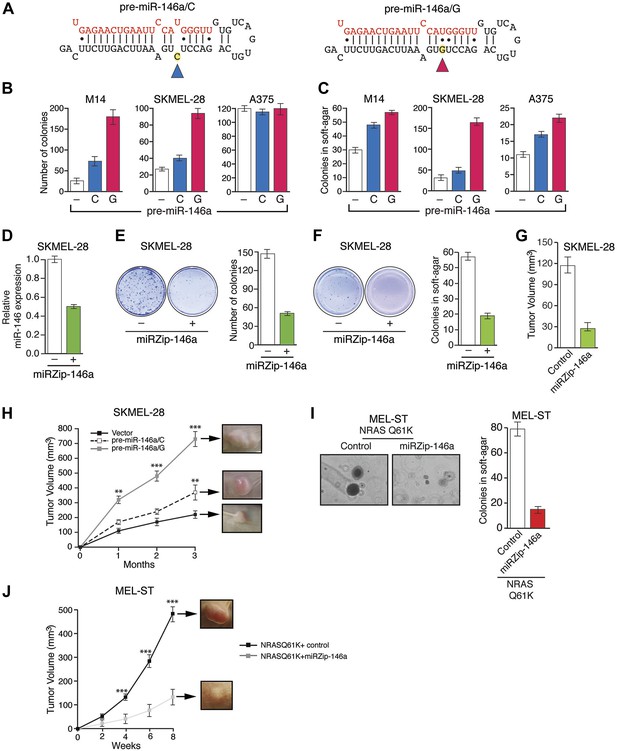
Oncogenic activity of pre-miR-146a/C and pre-miR-146a/G.
(A) Schematic representation of pre-miR-146a/C and pre-miR-146a/G sequences. (B and C) Number of colonies formed in liquid (B) or soft-agar (C) by M14, SKMEL-28 or A375 melanoma cells expressing pre-miR-146a harboring a C or G at position 40, as compared to the Vector (−) control. Colonies were counted after 2 weeks (B) or 4 weeks (C) of growth. (D) qRT-PCR analysis of miR-146a expression in SKMEL-28 cells infected with miRZip-146a (+) or an empty vector (−). (E and F) Number of colonies formed in liquid (E) or soft-agar (F) by SKMEL-28 expressing miRZip-146a (+) or the vector (−) control. Colonies were counted after 2 weeks (E) or 4 weeks (F) of growth. (G) Average tumor volumes at 1 month time points for mice injected with SKMEL-28 expressing a control miRZip or miRZip-146a. (H) Average volume of tumors formed by SKMEL-28 cells (2.5 × 106) expressing pre-miR-146a/C or pre-miR-146a/G, as compared to the Vector control, injected subcutaneously into the flanks of nude mice (n = 5). (I) Representative images (left) and colony number in soft-agar (right) for the NRASQ61K transformed MEL-ST cells that express an empty vector or miRZip-146a. (J) Average tumor volumes at the indicated time points for mice injected with NRASQ61K transformed MEL-ST cells expressing a control miRZip or miRZip-146a.
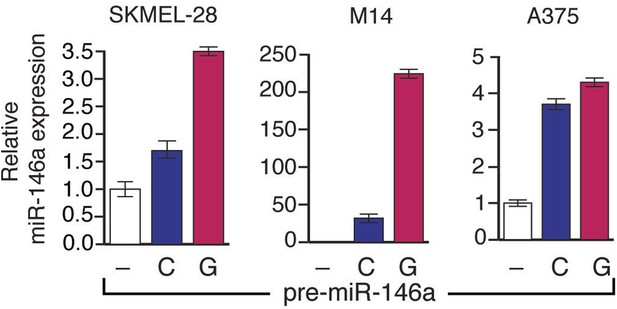
Analysis of miR-146a expression in SKMEL-28, A375 and M14 cells expressing either pre-miR-146a/G or pre-miR-146a/C construct.
SKMEL-28, M14 and A375 cells stably expressing pre-miR-146a/C (C) or pre-miR-146a/G (G) or an empty vector (−) were analyzed for miR-146a expression by qRT-PCR.
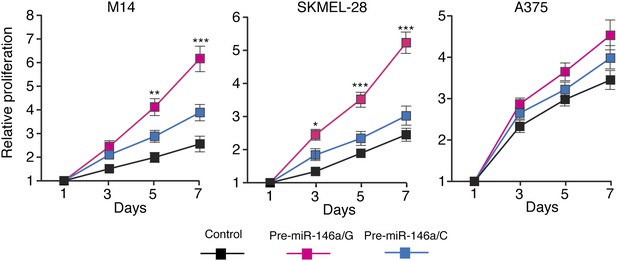
pre-miR-146a/G promotes proliferation of melanoma cells more effective than pre-miR-146a/C.
M14, SKMEL-28 and A375 cells stably expressing pre-miR-146a/C (blue) or pre-miR-146a/G (red) or an empty vector (black) were analyzed for proliferation at indicated days. Relative proliferation is plotted. *, ** and *** represents p values <0.01, <0.001 and <0.0001 respectively.
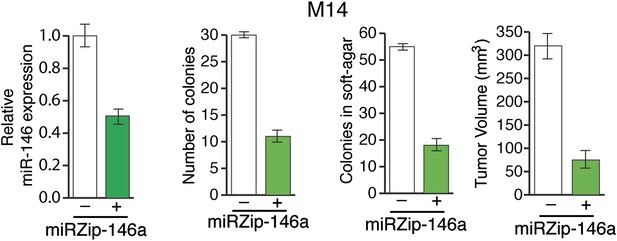
Inhibition of miR-146a expression blocks the proliferation and anchorage-independent growth of M14 cells.
M14 cells either expressing a control miRZip vector or miRZip-146a were analyzed for miR-146a expression, colony formation, or growth in soft-agar or tumor formation in mice. Tumor volume at 1 month timepoint is plotted.
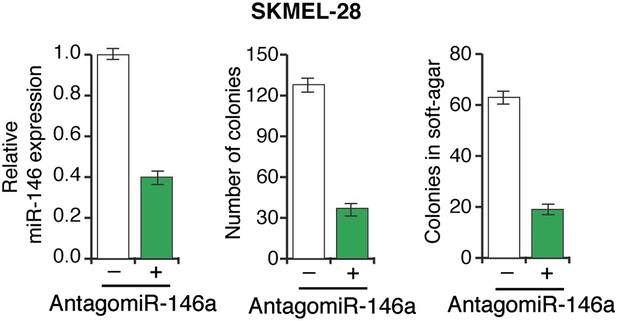
Inhibition of miR-146a expression blocks the proliferation and anchorage-independent growth of SKMEL-28 cells.
SKMEL-28 cells either expressing a scrambled LNA-antagomiR or LNA-based miR146a antagomiR were analyzed for miR-146a expression (left), colony formation (middle) or growth in soft-agar (right).
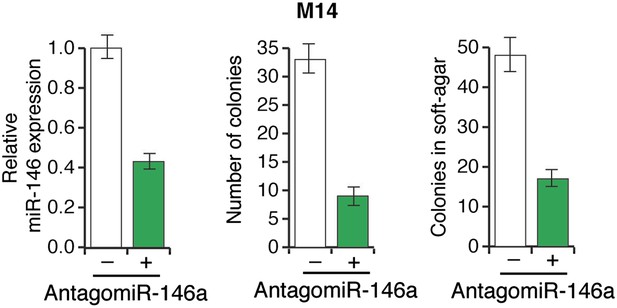
Inhibition of miR-146a expression blocks the proliferation and anchorage-independent growth of M14 cells.
M14 cells either expressing a scrambled LNA-antagomiR or LNA-based miR146a antagomiR were analyzed for miR-146a expression (left), colony formation (middle) or growth in soft-agar (right).
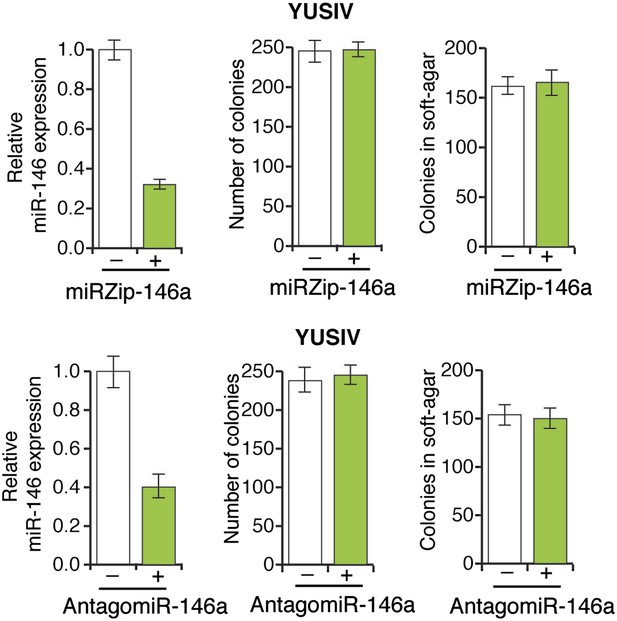
Inhibition of miR-146a expression does not block the proliferation and anchorage-independent growth of YUSIV cells.
(Top panel) YUSIV cells either expressing a control miR-ZIP vector or miR-Zip-miR146a were analyzed for miR-146a expression (left), colony formation (middle) or growth in soft-agar (right). (Bottom Panel) YUSIV cells either expressing a control LNA-based control antagomiR or LNA-based miR-146a antagomiR were analyzed for miR-146a expression (left), colony formation (middle) or growth in soft-agar (right).
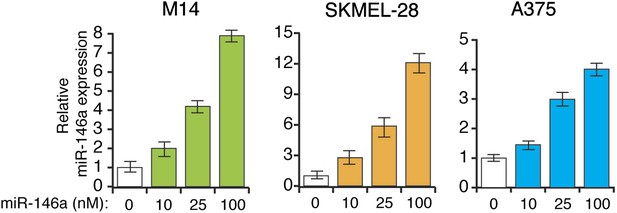
Analysis of miR-146a expression in indicated melanoma cell lines transfected with increasing concentration of synthetic miR-146a.
qRT-PCR analysis of miR-146a expression in indicated melanoma cell lines.
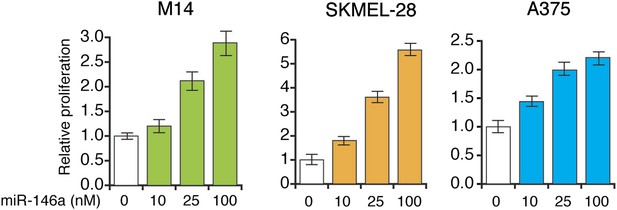
miR-146a enhances the growth of melanoma cell lines in a concentration dependent manner.
Relative proliferation of indicated melanoma cell lines 5 days after the transfection of synthetic mature miR-146a at indicated concentrations.
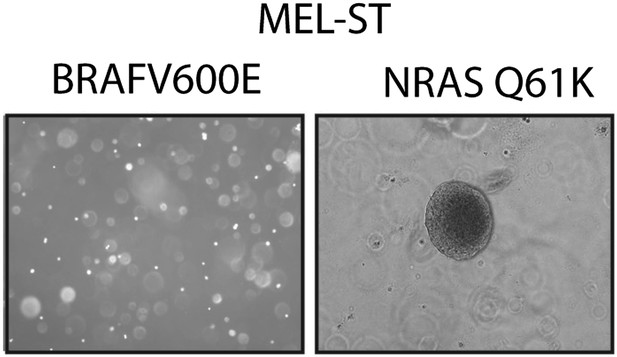
NRASQ61K is sufficient to transform MEL-ST cells.
Representative images of soft-agar colonies formed by MEL-ST cells expressing BRAFV600E. NRASQ61K was used as a positive control.
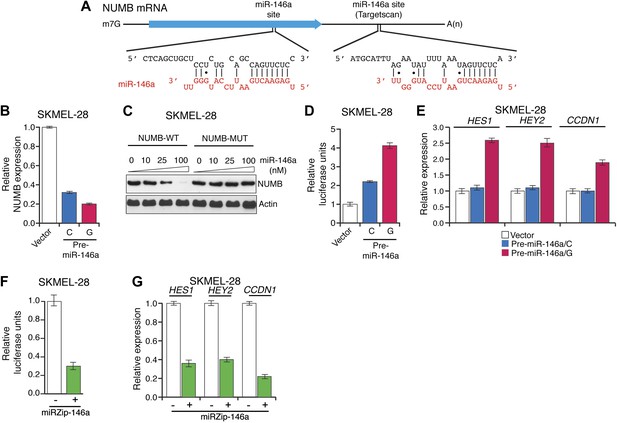
Downregulation of NUMB and activation of NOTCH signaling by pre-miR-146a/C and pre-miR-146a/G.
(A) Schematic representation of the NUMB mRNA and potential miR-146a target sites in the coding region (blue arrow) and 3′-UTR. (B) qRT-PCR analysis of NUMB mRNA levels in SKMEL-28 cells expressing the indicated pre-miR-146a allele relative to the Vector control. (C) Western blot of NUMB expression in SKMEL-28 cells transfected with either wild-type NUMB (NUMB-WT) or miR-146a-resistant NUMB (NUMB-MUT) and increasing amount of synthetic miR-146a. (D) Dual luciferase assay using a CSL-Luciferase reporter to measure NOTCH activity in SKMEL-28 cells expressing the indicated pre-miR-146a alleles relative to the Vector control. (E) qRT-PCR analysis of the NOTCH targets HES1, HEY2 and CCDN1 mRNA in indicated samples. Actin mRNA was used as an internal control. (F) Dual luciferase assay using a CSL-Luciferase reporter to measure NOTCH activity in SKMEL-28 cells expressing miRZip-146a relative to the Vector control. (G) qRT-PCR analysis of HES1, HEY2 and CCDN1 mRNA in indicated samples. Actin mRNA was used as a loading control.
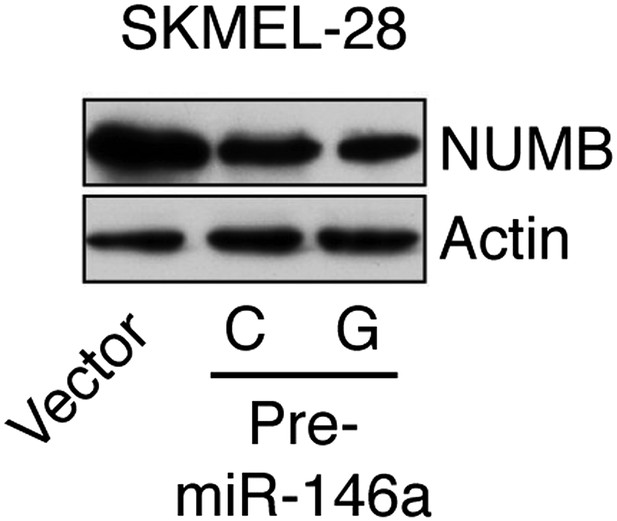
Ectopic expression of miR-146a downregulates NUMB protein expression.
Immunoblot analysis of NUMB protein in SKMEL-28 cells stably transduced with empty vector, pre-miR-146a/C or pre-miR-146a/G.
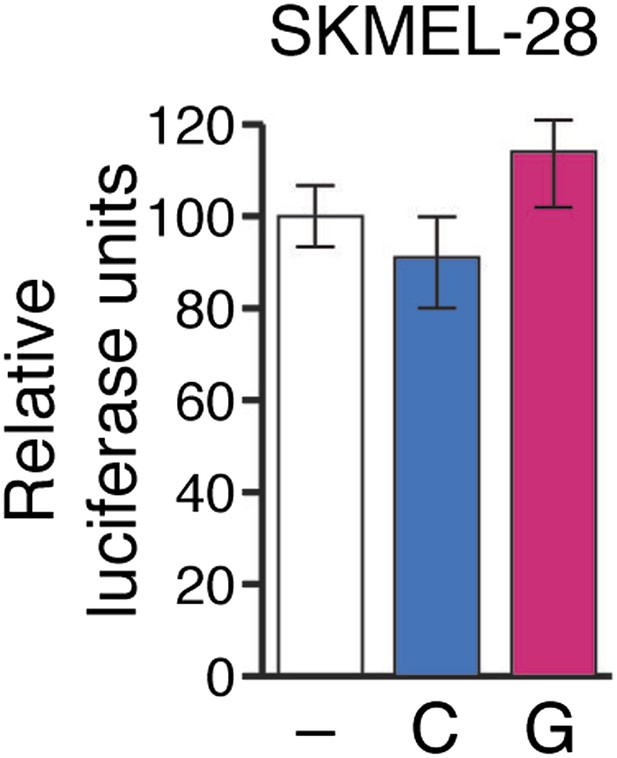
Relative luciferase activity of NUMB 3′UTR luciferase construct in SKMEL-28 cells expressing an empty vector (−), pre-miR-146a/C (C) or pre-miR-146a/G (G).
https://doi.org/10.7554/eLife.01460.026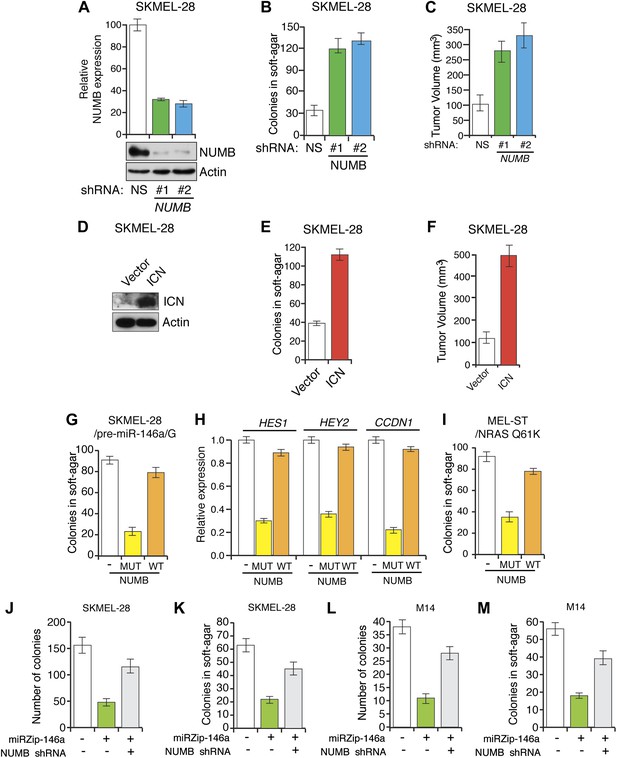
miR-146 oncogenic activity depends on the activation of the NOTCH signaling through downregulation of the tumor suppressor NUMB.
(A) qRT-PCR analysis (top) and immunoblot (bottom) of NUMB expression levels in SKMEL-28 cells infected with two different shRNAs against NUMB relative to the control non-silencing shRNA (NS). (B) Number of colonies in soft-agar of SKMEL-28 cells expressing NUMB shRNAs relative to the control non-silencing shRNA (NS). (C) Average volume of tumors formed by SKMEL-28 cells (2.5 × 106) expressing NUMB shRNAs, relative to the non-silencing shRNA (NS shRNA) control, injected subcutaneously into the flanks of nude mice (n = 5). (D and E) Immunoblot of NOTCH (D) and colony formation assay (E) of SKMEL-28 cells stably transduced with the activated intracellular NOTCH domain (ICN) or empty vector. (F) Average volume of tumors formed by SKMEL-28 cells expressing the activated intracellular NOTCH domain (ICN) relative to vector control. 2.5 × 106 cells were injected subcutaneously into the flank of nude mice (n = 5). (G) Colony formation in soft-agar of SKMEL-28 cells expressing pre-miR-146a/G and transfected with either an empty vector, NUMB wild-type (WT) or an miR-146a-resistant NUMB (MUT). (H) qRT-PCR analysis of HES1, HEY2 and CCDN1 mRNA in indicated samples. Actin was used as an internal control. (I) Colony formation in soft-agar of MEL-ST/NRASQ61K transfected with either an empty vector, NUMB wild-type (WT) or miR-146a-resistant NUMB (MUT). (J and K) Number of colonies formed in liquid (J) or soft-agar (K) by SKMEL-28 expressing miRZip-146a (+) or the control miRZip (−) that either express a non-silencing shRNA or an shRNA against NUMB. Colonies were counted after 2 weeks (J) or 4 weeks (K) of growth. (L and M) Number of colonies formed in liquid (L) or soft-agar (M) by M14 expressing miRZip-146a (+) or the control miRZip (−) that either express a non-silencing shRNA or an shRNA against NUMB. Colonies were counted after 2 weeks (L) or 4 weeks (M) of growth.
Inhibition of NUMB expression promotes growth of SKMEL-28 cells.
Colony formation assay of SKMEL-28 cells expressing two different NUMB shRNAs relative to a non-silencing (NS) shRNA.
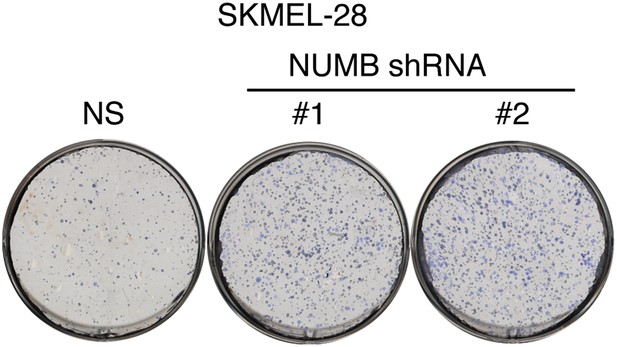
Inhibition of NUMB promotes proliferation of melanoma cells.
Proliferation assay of SKMEL-28 cells expressing two different NUMB shRNAs relative to a non-silencing (NS) shRNA.
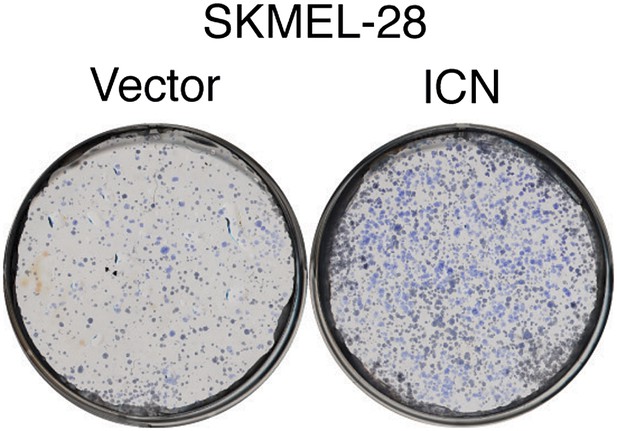
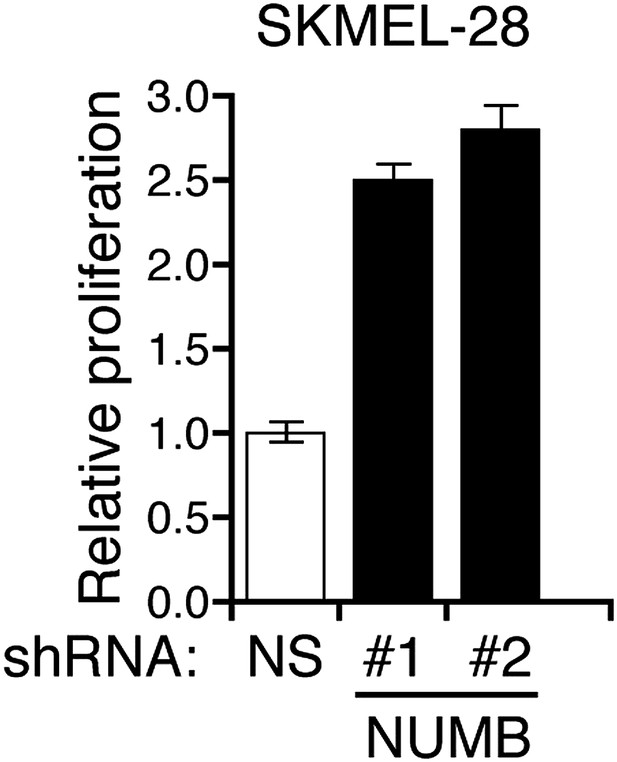
Ectopic expression of activated Notch promotes the growth of melanoma cells.
Colony formation assay of SKMEL-28 cells stably transduced with the activated intracellular NOTCH domain (ICN) or empty vector.
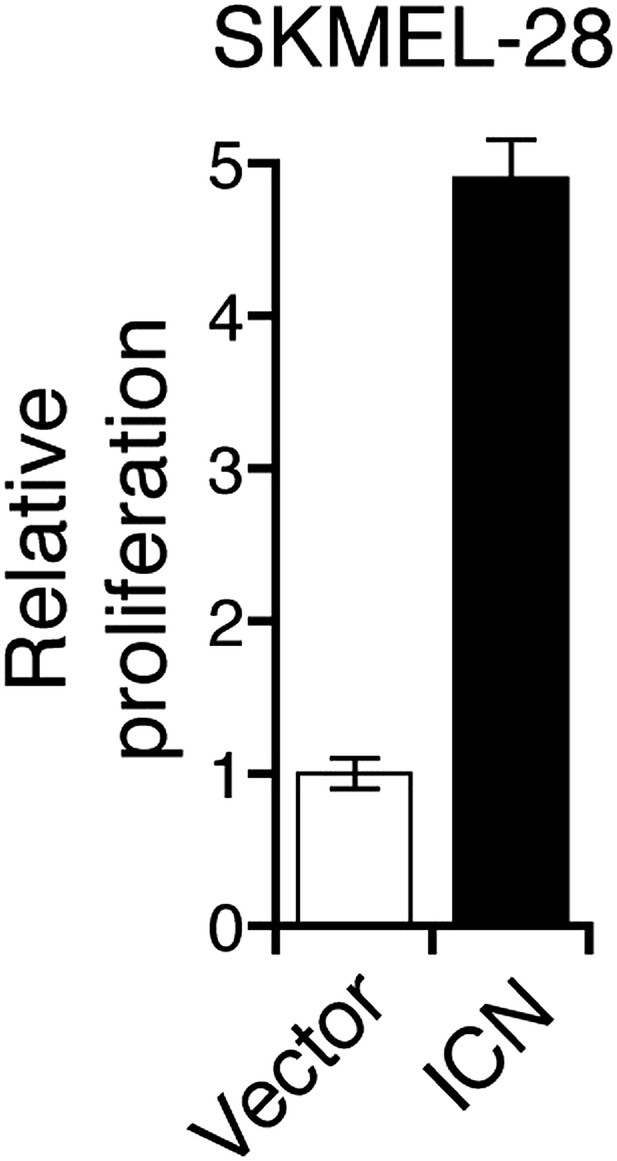
Ectopic expression of intracellular Notch promotes proliferation of melanoma cells.
Proliferation rate of SKMEL-28 cells stably expressing Intracellular NOTCH (ICN) relative to cells with empty vector. Proliferation rate was measured after 72 hr of growth using a colorimetric MTT assay.
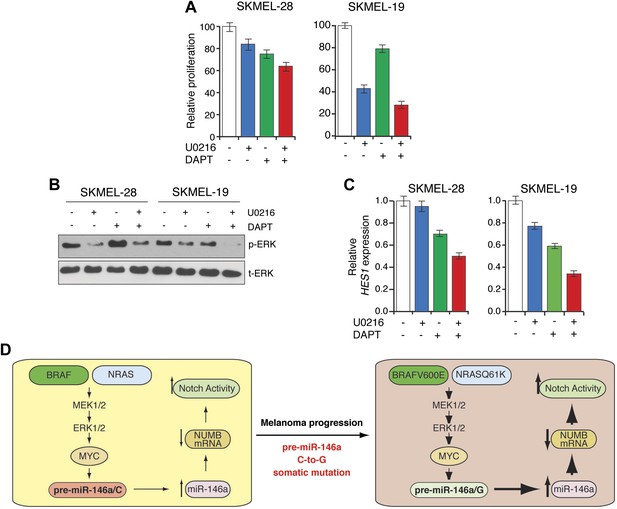
Synergistic melanoma growth inhibition by simultaneous blockage of Notch and BRAF signaling.
(A) MTT proliferation assay. (B) Immunoblot analysis of phosphorylated (p-) ERK and total (t-) ERK and (C) qRT-PCR analysis of NOTCH target HES1 in SKMEL-28 and SKMEL-19 cells left untreated or treated with U0216, DAPT or combination of both drugs. (D) Model.
Tables
Sequence at position 40 of pre-miR-146a in primary human melanocytes, human melanoma cell lines and clinical samples
| Human melanoma cell lines | |
|---|---|
| Cell Line | |
| Melanocytes-1 | CC |
| Melanocytes-2 | CC |
| Yale SPORE melanocytes | CG |
| WM3918 | CG |
| YUHEF | CG |
| YUVON | CG |
| YUSIV | GG |
| YUROB | GG |
| YUROL | CG |
| MeWo | GG |
| UCC257 | CG |
| SKMEL28 | CG |
| A375 | GG |
| SKMEL5 | GG |
| M14 | CG |
| SKMEL19 | GG |
| YULAC | GG |
| YUGEN8 | GG |
| YUSAC2 | GG |
| YURIF | CG |
| SKMEL103 | CG |
| Matched clinical melanoma samples (Nevus/Primary) | ||
|---|---|---|
| Sample | Nevus (Type)* | Primary Melanoma |
| 1 | CG (IM) | CG |
| 2 | CC (C) | CG |
| 3 | CG (C) | GG |
| 4 | CG (C) | GG |
| 5 | CG (LJ) | CG |
| 6 | CG (IM) | CG |
| 7 | CG (IM) | CG |
| 8 | CG (LJ) | CG |
| 9 | CG (IM) | CG |
| 10 | CC (C) | CG |
| Matched Clinical Melanoma Samples (Primary/Metastatic) | ||
|---|---|---|
| Sample | Primary Melanoma | Metastatic Melanoma |
| 1 | CG | GG |
| 2 | CG | CG |
| 3 | CG | GG |
| 4 | CG | CG |
| 5 | CC | GG |
| 6 | CG | GG |
| 7 | CG | GG |
| 8 | CC | CC |
| 9 | CG | CG |
| 10 | CG | CG |
| 11 | CG | CG |
| 12 | CG | GG |
| 13 | CG | GG |
| 14 | CG | GG |
-
*
IM, Intradermal melanocytic nevus. C, Compound nevus. LJ, Lentigenous juctional nevus. Samples highlighted in gray indicate C-to-G mutation during melanoma progression.
Additional files
-
Supplementary file 1
(A) List of genes down-regulated by both pre-miR-146a/C and pre-miR-146a/G with miR-146a seed sequences (by TargetScan). (B) Genotyping of patient samples for BRAF and NRAS mutations. (C) Mutant BRAF, mutant NRAS and miR-146a C and G allele frequencies. (D) Melanoma SNP dataset analyses for pre-miR-146a/C and pre-miR-146a/G frequencies. (E) Primer sequences for qRT-PCR analysis; clone ID and catalog numbers for shRNAs (Open Biosystems); antibodies used; source and concentration of chemical inhibitors used.
- https://doi.org/10.7554/eLife.01460.034





