A serine sensor for multicellularity in a bacterium
Figures
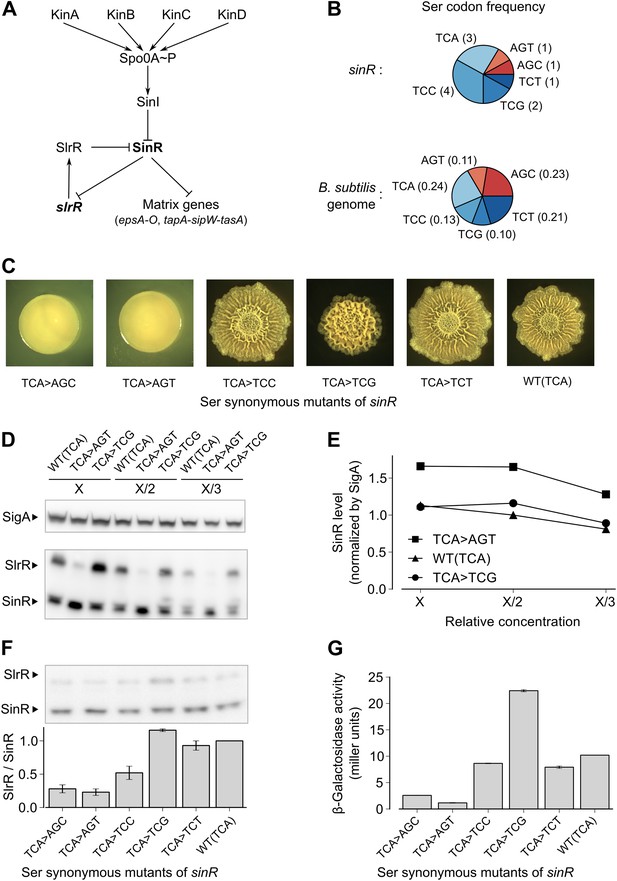
Switching synonymous serine codons in sinR affects biofilm formation.
(A) Regulatory circuit controlling biofilm formation in B. subtilis. (B) Top: Serine codon usage in the sinR coding sequence. Number within parenthesis indicates the frequency of the corresponding codon in sinR. Bottom: Average serine codon usage across 4153 protein-coding sequences in the B. subtilis genome. Number within parenthesis indicates the relative frequency of each codon in the genome. (C) Colony morphology for the wild-type strain and the indicated sinR synonymous variants grown on solid biofilm-inducing medium. Three TCA codons in the wild-type sequence of sinR were switched to each of the other five serine codons. The wild-type (WT) sinR sequence was replaced by the sinR synonymous mutant at the native sinR locus of the strain 3610. (D) SinR protein level during entry into biofilm formation (OD600 = 2) measured using an anti-SinR antibody that also cross-reacts with SlrR, a protein that is 85% identical to SinR. Western blot against the RNA polymerase subunit SigA was used as the loading control. Whole cell lysates were loaded at different dilutions (indicated as X, X/2, and X/3). (E) Densitometry of SinR bands in (D) after normalization by SigA. (F) Top panel: Western blot against SinR and SlrR using anti-SinR antibody. Bottom panel: Densitometry ratio of the SlrR and SinR bands in the top panel. Error bars represent standard error over three replicate Western blots. The SlrR/SinR ratio for each blot was normalized such that the wild-type strain had a ratio of 1. (G) Matrix gene expression monitored using a PepsA–lacZ transcriptional reporter inserted at the chromosomal amyE locus. β-galactosidase activity was measured at OD600 = 2 in liquid biofilm-inducing medium. Error bars represent standard error of three measurements.
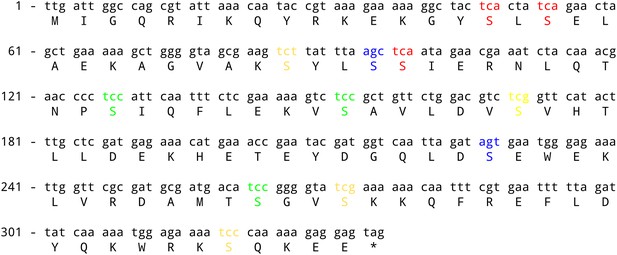
sinR coding sequence.
The three TCA codons (switched in Figure 1) are highlighted in red. The three TCC codons and the two AGC/AGT codons (switched in Figure 1—figure supplement 2) are highlighted in green and blue respectively. The remaining serine codons are shown in yellow.
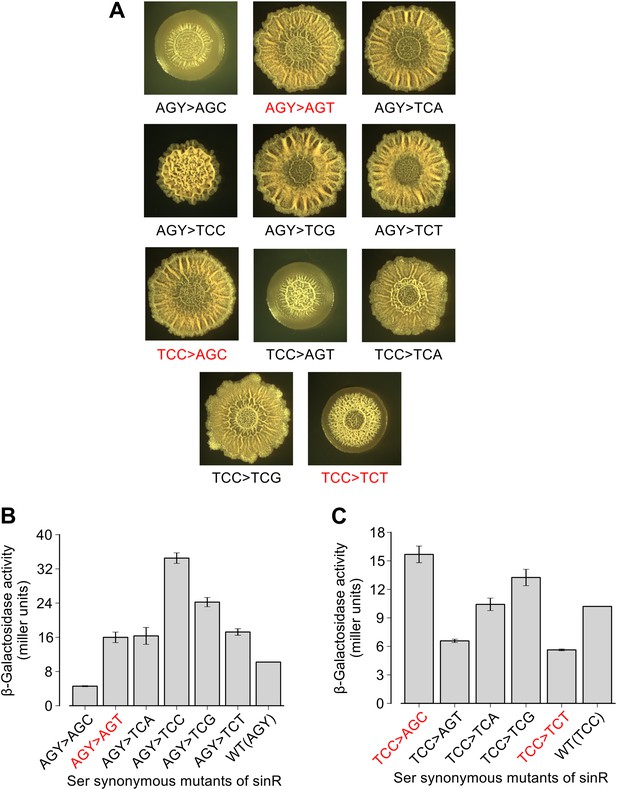
Effect of TCC and AGC/AGT synonymous substitutions in the sinR gene on colony morphology and biofilm reporter activity.
(A) Colony morphology for the indicated sinR synonymous variants grown on solid biofilm-inducing medium. Either three TCC codons or two AGC/AGT (AGY) codons in the wild-type sequence of sinR were switched to remaining serine synonymous codons. The wild-type (WT) sinR sequence was replaced by the sinR synonymous variant at the native sinR locus of the strain 3610. Colony morphology of the wild-type strain is shown in Figure 1. (B and C) Matrix gene expression monitored using a PepsA–lacZ transcriptional reporter inserted at the chromosomal amyE locus. Strains were grown in liquid biofilm-inducing medium and β-galactosidase activity was measured at an OD600 = 2. Error bars represent standard error of three measurements. The synonymous variants highlighted in red do not follow the hierarchy between TCN and AGC/AGT codons seen for the six TCA synonymous variants in Figure 1.
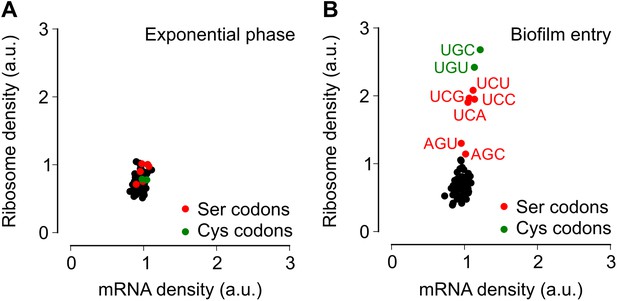
Entry into biofilm formation is accompanied by codon-specific increase in ribosome density.
Genome-wide median ribosome density and total mRNA density at 61 sense codons (excluding start and stop codons) (A) during exponential phase growth (OD600 = 0.6), and (B) during stationary phase when biofilm formation is induced (OD600 = 1.4). The six serine (red) and two cysteine (green) codons are highlighted. Genome-wide ribosome density and total mRNA density were measured by deep-sequencing of ribosome-protected mRNA fragments and total mRNA fragments respectively, of a B. subtilis 3610 derivative (ΔepsH) grown in liquid biofilm-inducing medium.
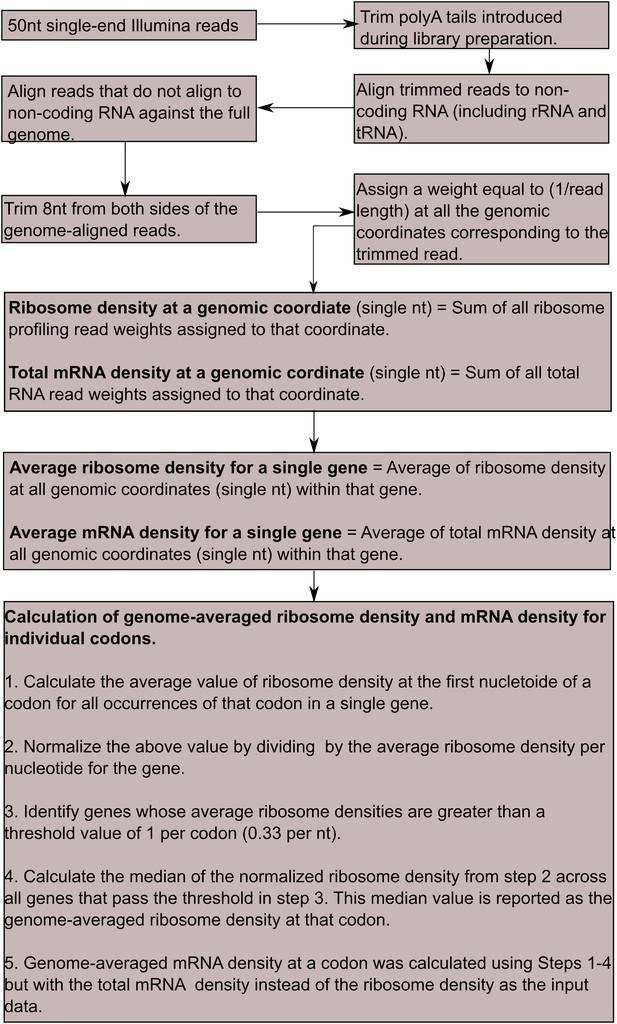
Computational workflow for deep-sequencing data analysis.
All steps outlined here were performed in Bash and Python programming languages. For further details on individual steps, see ‘Materials and methods’.
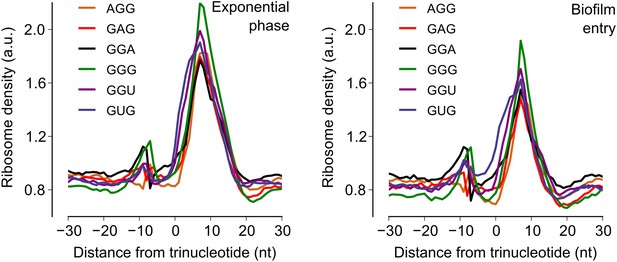
Increase in ribosome density downstream of Shine-Dalgarno-like trinucleotide sequences.
Median ribosome density across all protein coding sequences was computed for the 60 nt region around each of six Shine-Dalgarno-like trinucleotide sequences (Li et al., 2012) for the exponential phase sample (left-hand panel) and the biofilm entry sample (right-hand panel).
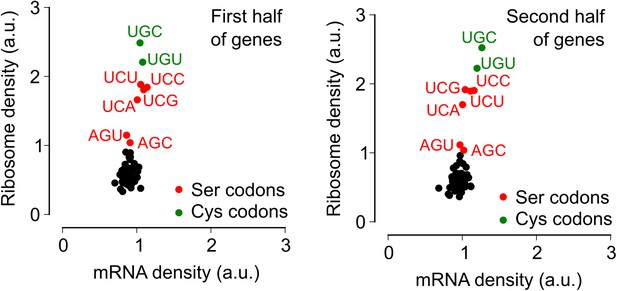
Context independence of ribosome and mRNA densities during biofilm formation.
Each gene was conceptually divided into two equal halves and the ribosome density and mRNA density was computed separately for codons located either in the first half (left-hand panel) or in the second half (right-hand panel) of each gene. All other analysis steps were identical to those in Figure 2.
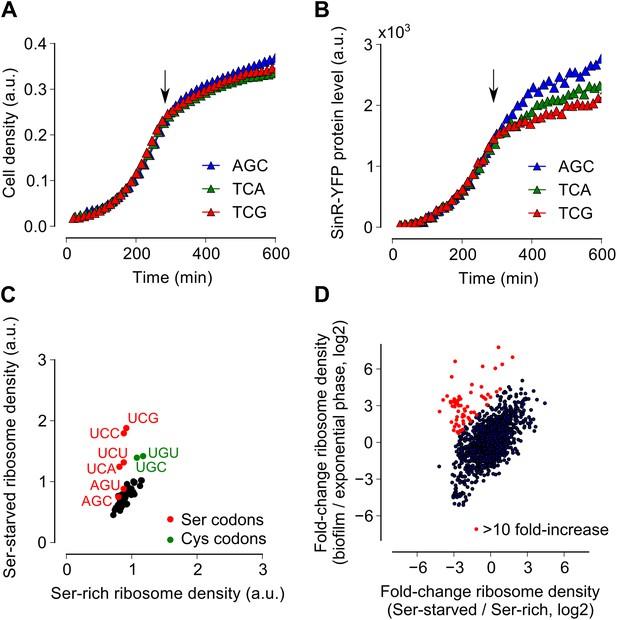
Serine starvation reduces translation speed and inhibits SinR synthesis in a codon-specific manner.
(A and B) Three sinR synonymous variants were synthesized with 10 serine codons switched to AGC, TCA or TCG. The variants were expressed as SinR-YFP fusions from the amyE locus under the control of a lac promoter in a 3610-ΔserA serine auxotroph strain growing in serine-limited medium. Black arrow around 300 min indicates the onset of serine starvation caused by depletion of exogenously-added serine in the growth medium. Cell density (A) and the corresponding SinR-YFP protein level (B) were monitored using a 96-well spectrophotometer. (C) Genome-wide median ribosome density for 61 sense codons (excluding start and stop codons) during serine starvation (vertical axis) and serine-rich growth (horizontal axis) of a serine auxotrophic strain. (D) Fold-change in average ribosome density for individual genes upon biofilm entry (vertical axis) or serine starvation (horizontal axis). Genes that were preferentially up-regulated at least 10-fold upon biofilm entry in comparison to serine starvation are highlighted in red (68 genes, Table 1). Only genes with a minimum of 100 ribosome profiling reads in at least one of the samples were included in this analysis (1724 genes) and the reported log2 fold-changes are median-subtracted values across this gene set.
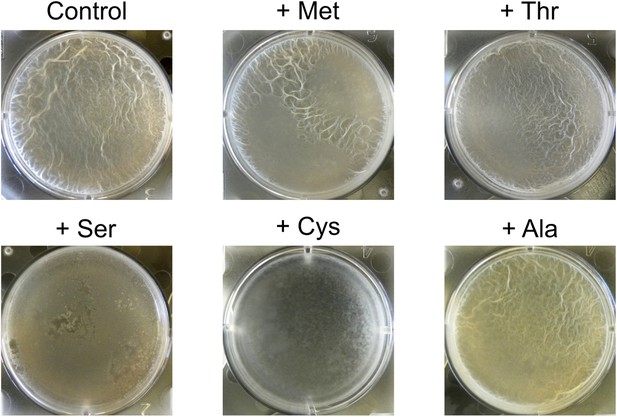
Addition of excess serine or cysteine blocks pellicle formation by B. subtilis.
Single amino acids were added at 300 µg ml−1 to liquid MSgg medium. Biofilm formation of 3610 was assayed visually by pellicle formation at the air-liquid interface 48 hr after inoculation. Serine and cysteine were found to block pellicle formation out of all 20 amino acids tested.
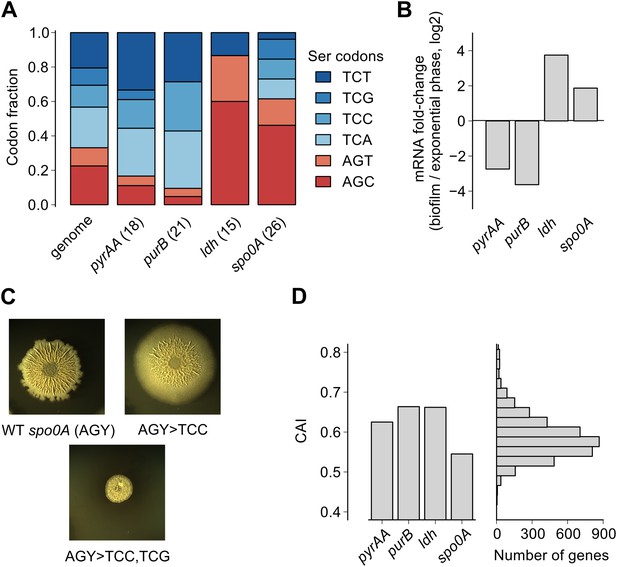
Serine codon bias of biofilm-regulated genes reflects their expression under serine starvation.
(A) Relative serine codon fraction in genes for nucleotide biosynthesis (pyrAA, purB), lactate dehydrogenase (ldh) and a sporulation regulator (spo0A). Numbers in parentheses indicate the number of serine codons in each gene. Relative fraction of serine codons across the B. subtilis genome is shown for comparison. (B) Fold-change (expressed in log2 units) in average ribosome density upon biofilm entry for the four genes shown in A. (C) Colony morphology of a wild-type strain and two spo0A synonymous variants grown on solid biofilm-inducing medium. Seven AGC/AGT codons in wild-type spo0A were replaced by either 7 TCC codons or 3 TCC and 4 TCG codons and inserted at the chromosomal spo0A locus. Both the wild-type spo0A and the synonymous spo0A variants were inserted with a downstream selection marker. (D) Left: Codon Adaptation Index (CAI) for the four genes shown in A. Right: Distribution of CAI values for 4153 protein-coding sequences of B. subtilis.
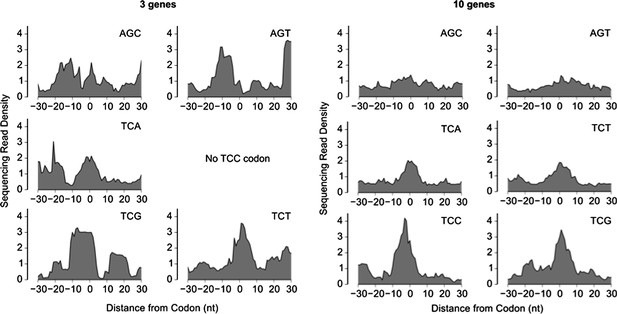
Sequencing read density around six serine codons, calculated as a median over the 3 genes with the highest ribosome density (left) or the 10 genes with the highest ribosome density (right). The sequencing read density for each gene was normalized by the total number of sequencing reads for that gene (same procedure as in Figure 2).
Tables
B. subtilis genes that have greater than 10-fold difference in expression ratio between biofilm formation and serine starvation
| Gene | A | B | C | D | E | F | Function |
|---|---|---|---|---|---|---|---|
| albA | 9.35 | −0.69 | 32 | 10129 | 175 | 144 | antilisterial bacteriocin (subtilosin) production protein |
| alsD | 6.38 | 0.95 | 34 | 1384 | 53 | 135 | alpha-acetolactate decarboxylase |
| alsS | 6.96 | 1.79 | 76 | 4613 | 86 | 396 | acetolactate synthase |
| cah | 2.08 | −2.74 | 2531 | 5254 | 1486 | 295 | S-deacylase |
| ctc | 3.78 | −0.73 | 327 | 2206 | 322 | 257 | 50S ribosomal protein L25 |
| cydA | 5.36 | −3.18 | 234 | 4720 | 408 | 60 | cytochrome bd ubiquinol oxidase subunit I |
| cydB | 9.6 | −1.88 | 9 | 3374 | 129 | 46 | cytochrome bd ubiquinol oxidase subunit II |
| cysK | 1.52 | −2.21 | 15760 | 22120 | 8991 | 2580 | cysteine synthase |
| gcvPA | 0.97 | −2.38 | 1500 | 1440 | 1001 | 255 | glycine dehydrogenase subunit 1 |
| gcvPB | 1.05 | −2.28 | 2029 | 2057 | 1224 | 335 | glycine dehydrogenase subunit 2 |
| gspA | 4.54 | −0.75 | 111 | 1261 | 135 | 106 | glycosyl transferase (general stress protein) |
| iseA | 2.24 | −1.14 | 1117 | 2579 | 2521 | 1516 | inhibitor of cell-separation enzymes |
| lctP | 6.62 | −2.9 | 62 | 3009 | 247 | 44 | L-lactate permease |
| ldh | 3.47 | −3.88 | 2810 | 15,254 | 1732 | 157 | L-lactate dehydrogenase |
| maeN | 4.08 | −1.61 | 148 | 1226 | 347 | 151 | Na+/malate symporter |
| mccA | 3.23 | −2.72 | 759 | 3494 | 386 | 78 | cystathionine beta-synthase |
| metE | 2.72 | −2.13 | 33,027 | 106473 | 32,231 | 9760 | 5-methyltetrahydropteroyltriglutamate/homocysteine S-methyltransferase |
| mgsR | 4.72 | −1.53 | 163 | 2099 | 202 | 93 | stress transcriptional regulator |
| mtlA | 3.92 | −0.22 | 126 | 934 | 104 | 119 | PTS system mannitol-specific transporter subunit IICB |
| mtnA | 1.84 | −2.79 | 2522 | 4425 | 3501 | 671 | methylthioribose-1-phosphate isomerase |
| mtnD | 1.72 | −1.63 | 4095 | 6600 | 3182 | 1361 | acireductone dioxygenase |
| mtnK | 2.54 | −2.64 | 4290 | 12,196 | 5866 | 1250 | methylthioribose kinase |
| nasD | 6.21 | −0.65 | 217 | 7872 | 635 | 536 | assimilatory nitrite reductase subunit |
| nasE | 6.05 | 0.38 | 34 | 1106 | 63 | 109 | assimilatory nitrite reductase subunit |
| rbfK | 3.74 | −2.57 | 1329 | 8681 | 434 | 97 | RNA-binding riboflavin kinase |
| sboA | 7.77 | 0.64 | 121 | 12,895 | 256 | 530 | subtilosin A |
| sboX | 8.26 | 0.19 | 21 | 3128 | 76 | 115 | bacteriocin-like product |
| ssuA | 2.09 | −2.3 | 3691 | 7682 | 877 | 236 | aliphatic sulfonate ABC transporter binding lipoprotein |
| ssuB | 2.57 | −2.09 | 2416 | 7004 | 779 | 242 | aliphatic sulfonate ABC transporter ATP-binding protein |
| ssuC | 2.17 | −2.16 | 3784 | 8362 | 692 | 206 | aliphatic sulfonate ABC transporter permease |
| ssuD | 2.2 | −2.19 | 12,961 | 29,135 | 2812 | 817 | alkanesulfonate monooxygenase |
| tcyJ | 3.25 | −3.26 | 1046 | 4856 | 427 | 59 | sulfur-containing amino acid ABC transporter binding lipoprotein |
| tcyK | 3.79 | −3.55 | 2815 | 19,087 | 1095 | 124 | sulfur-containing amino acid ABC transporter binding lipoprotein |
| tcyL | 3.33 | −3.05 | 855 | 4223 | 387 | 62 | sulfur-containing amino acid ABC transporter permease |
| tcyM | 3.54 | −2.97 | 1859 | 10,556 | 581 | 99 | sulfur-containing amino acid ABC transporter permease |
| tcyN | 3.38 | −2.74 | 3363 | 17,149 | 1119 | 223 | sulfur-containing amino acid ABC transporter ATP-binding protein |
| ureA | 4.36 | 0.78 | 124 | 1252 | 77 | 176 | urease subunit gamma |
| ycgL | 0.99 | −3.01 | 512 | 500 | 278 | 46 | hypothetical protein |
| ycgM | 2.46 | −1.35 | 39 | 105 | 178 | 93 | proline oxidase |
| ycgN | 2.45 | −1.6 | 928 | 2475 | 1267 | 556 | 1-pyrroline-5-carboxylate dehydrogenase |
| ycnJ | 0.75 | −2.63 | 168 | 138 | 121 | 26 | copper import protein |
| ydaG | 4.14 | 0.49 | 70 | 604 | 80 | 150 | general stress protein |
| ydbL | 3.07 | −0.31 | 294 | 1210 | 130 | 139 | hypothetical protein |
| yeaA | 1.34 | −2.59 | 112 | 138 | 139 | 31 | hypothetical protein |
| yezD | 2.52 | −4.17 | 145 | 406 | 340 | 25 | hypothetical protein |
| yitJ | 3.01 | −2.43 | 2785 | 10,990 | 4687 | 1153 | bifunctional homocysteine S-methyltransferase/5,10-methylenetetrahydrofolate reductase |
| yjbC | 3.67 | −0.57 | 104 | 647 | 248 | 222 | thiol oxidation management factor; acetyltransferase |
| yjnA | 1.5 | −2.81 | 972 | 1342 | 790 | 149 | hypothetical protein |
| yoaB | 2.28 | −2.26 | 2206 | 5244 | 2862 | 792 | negatively charged metabolite transporter |
| yoaC | 2.92 | −1.89 | 1200 | 4459 | 1436 | 513 | hydroxylated metabolite kinase |
| yrhB | 2.92 | −2.93 | 3806 | 14,096 | 1904 | 333 | cystathionine beta-lyase |
| yrrT | 2.97 | −3.12 | 546 | 2089 | 443 | 68 | AdoMet-dependent methyltransferase |
| ytlI | 1.66 | −3.2 | 206 | 318 | 141 | 20 | LysR family transcriptional regulator |
| ytmI | 3.34 | −3.27 | 3452 | 17,173 | 1640 | 226 | N-acetyltransferase |
| ytmO | 3.4 | −2.88 | 3866 | 20,007 | 1179 | 213 | monooxygenase |
| ytnI | 3 | −2.6 | 3522 | 13,770 | 867 | 189 | redoxin |
| ytnJ | 3.14 | −2.86 | 10,645 | 45,997 | 2495 | 456 | monooxygenase |
| ytnL | 3.56 | −2.45 | 1281 | 7371 | 354 | 86 | aminohydrolase |
| ytnM | 3.5 | −2.6 | 4542 | 25,202 | 1264 | 277 | transporter |
| yuaF | 1.89 | −1.74 | 87 | 157 | 158 | 63 | membrane integrity integral inner membrane protein |
| yvzB | 0.95 | −2.48 | 125 | 118 | 168 | 40 | flagellin |
| yxaL | 3.54 | −0.41 | 1069 | 6077 | 442 | 442 | membrane associated protein kinase |
| yxbB | 3.7 | −0.01 | 108 | 685 | 118 | 155 | S-adenosylmethionine-dependent methyltransferase |
| yxeK | 0.86 | −2.72 | 2702 | 2406 | 1073 | 216 | monooxygenase |
| yxeL | 1.29 | −2.94 | 437 | 525 | 202 | 35 | acetyltransferase |
| yxeM | 0.87 | −2.57 | 3003 | 2692 | 1047 | 233 | ABC transporter binding lipoprotein |
| yxeP | 1.75 | −2.24 | 2577 | 4246 | 736 | 207 | amidohydrolase |
| yxjH | 2.02 | −1.82 | 4162 | 8243 | 3811 | 1432 | methyl-tetrahydrofolate methyltransferase |
-
A—median-subtracted log2 fold-change: biofilm/exponential-phase, B—median-subtracted log2 fold-change: serine starvation/serine rich, C—raw counts: biofilm entry, D—raw counts: exponential phase, E—raw counts: serine rich, F—raw counts: serine starvation.
Additional files
-
Supplementary file 1
Lists of strains, plasmids, and primers.
- https://doi.org/10.7554/eLife.01501.014





