A lipid bound actin meshwork organizes liquid phase separation in model membranes
Figures
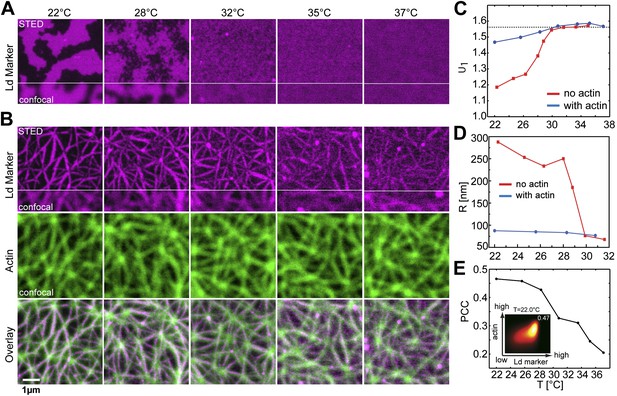
Binding of an actin network to ternary membranes dramatically affects lipid domain structure.
(A) In the absence of actin, our Mica supported model membrane revealed a phase transition at Tc ≈ 28°C, below which macroscopic Lo/Ld phase separation was observed. The membrane was stained with the Ld-marker DPPE-KK114 (magenta), and imaged by STED-microscopy with a lateral resolution of 70 nm. (B) The same membrane as in (A) but now in the presence of an actin network (green) bound to the membrane via a streptavidin linker. The Ld domains were strongly correlated to the actin network, as can be seen in the overlay, resulting in a meshwork like structure of Ld channels. This structure was stable even at temperatures above Tc. (C) Variation of the cumulant U1 of the images with temperature T in the absence of actin (red) and presence (blue). In the absence of actin, there is a transition toward a two-phase coexistence region at low temperature, as manifested by a pronounced drop of U1 toward unity. The transition occurs at Tc ≈ 28°C. In the presence of actin, no such transition is detected. (D) Typical domain size R as a function of T. In the absence of actin, there was pronounced domain coarsening below Tc. In the presence of actin, R was essentially temperature independent, and restricted to at most 90 nm. Note that R could not be measured for temperatures above T = 32°C as the contrast of the domains became too low. (E) Pearson correlation coefficient (PCC) between Ld domains and actin vs temperature. This coefficient measures the degree of correlation between Ld domains and the actin fibers. The correlation is largest at low temperature, but it persists at high temperature also, including the physiological temperature T = 37°C. Inset shows the graphical representation of the correlation between high intensities in the actin channel with high intensities in the lipid channel (ld phase). For another example of phase reorganization by actin binding see Figure 1—figure supplement 1. For actin meshwork binding to single component DOPC membranes see Figure 1—figure supplement 2.
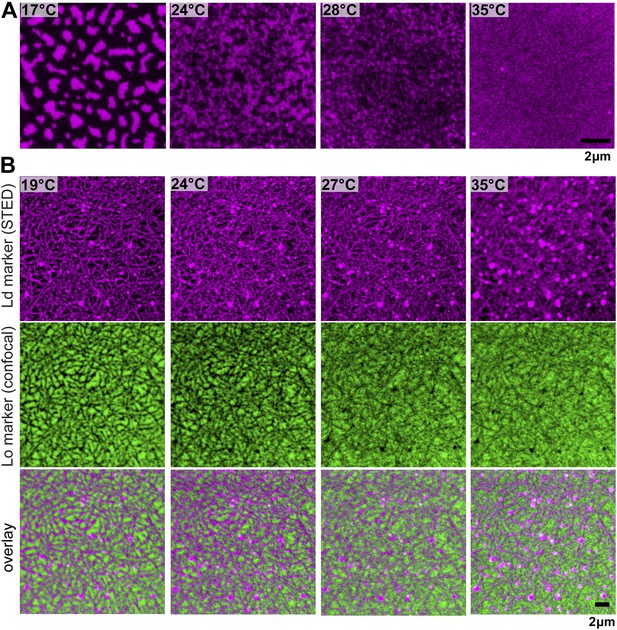
Phase organization by the actin-network.
Simultaneous confocal and STED images of a fluorescent lipid preferring the Lo (DSPE-PEG-Chromeo488, green, Lo marker) and Ld phase (DPPE-KK114, red, Ld marker), respectively. The membrane has the same composition as in Figure 1 without staining the actin network, confirming a negligible influence of the green-fluorescent phalloidin actin marker. (A) Without actin, the Ld marker reveals the typical phase separating behavior: below Tc domains coarsen, while above Tc the membrane is homogenous (Tc ≈ 28°C). (B) After actin binding to DOPE-biotin, Ld domains form a meshwork-like structure, similar to Figure 1B. The Lo domains form an inverse pattern, that is compartments separated by the Ld meshwork. As in Figure 1, the mosaic pattern persists above the phase transition temperature Tc of the membrane without actin.
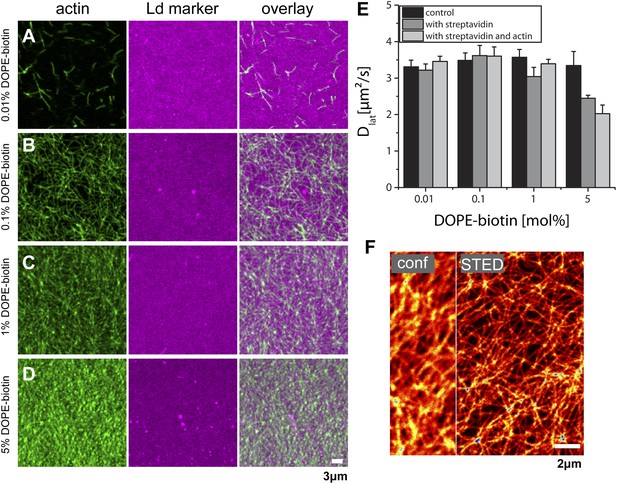
Actin-network density and lipid diffusion dependence on the concentration of biotinylated lipids. The data refer to a single lipid species (DOPC) supported bilayer at T = 22°C.
(A–D) Images of actin binding for increasing concentrations of DOPE-biotin (as marked on the left) using fluorescence markers phalloidin-488 for actin and DPPE-KK114 for the Ld phase. As expected, the actin-network density increases with the number of binding sites in the membrane. In contrast to ternary membranes (Figure 1), the Ld-marker distribution remains homogeneous. (E) Measurements of the lateral diffusion constant Dof DPPE-KK114 (Ld marker) determined with FCS. For increasing concentrations of DOPE-biotin, the diffusion constant was determined before the addition of streptavidin (control), after the addition of streptavidin (with streptavidin), and after the subsequent addition of actin (i.e., with streptavidin and actin). No change in diffusion was detected after actin binding for DOPE-biotin concentrations up to 1 mol%. Only for very high concentrations (5 mol%) could a decrease (∼30%) in lateral diffusion be detected. (F) STED image of the actin-network at a binding site concentration of 1 mol% DOPE-biotin (actin was stained with Alexa633-phalloidin). The network can be clearly resolved by STED. Compartment sizes are between 60 nm and 500 nm with a peak at 120 nm.
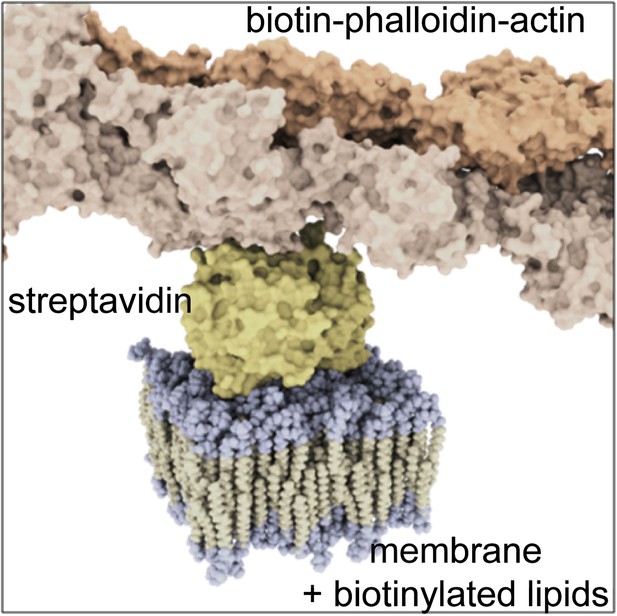
Molecular sketch of the assembled components in our model system.
https://doi.org/10.7554/eLife.01671.006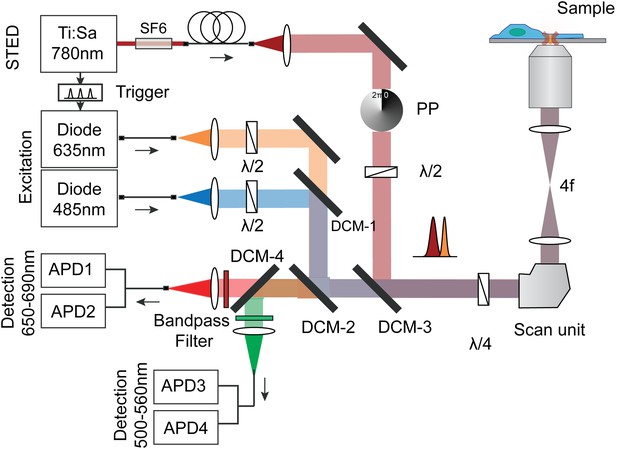
Optical setup for STED imaging and scanning FCS with pulsed excitation and pulsed STED laser and according beam paths (excitation orange/blue and STED dark red).
The repetition rates of the three lasers were synchronized using an electronic trigger box. The laser beams were overlaid using three dichroic mirrors and focused into the sample. The emitted fluorescence (red/green) from the sample was split with a dichroic mirror and cleaned using bandpass filters and detected using four fiber-coupled avalanche photon detectors (APD). Each fluorescence channel was imaged onto the input of a 50:50 splitting fiber, serving as the confocal pinhole. The polymeric phase plate (PP) induced a clockwise 2π-phase shift across the STED beam, which generated the doughnut shaped intensity distribution in the focal plane. Note, that the circular polarization of all laser beams is maintained by a combination of λ/2 and λ/4 waveplates. The STED laser was further sent through SF6 glass rods (SF6) and a 120 mlong polarization maintaining single-mode fiber (PMS) for stretching of the pulses and mode cleaning.
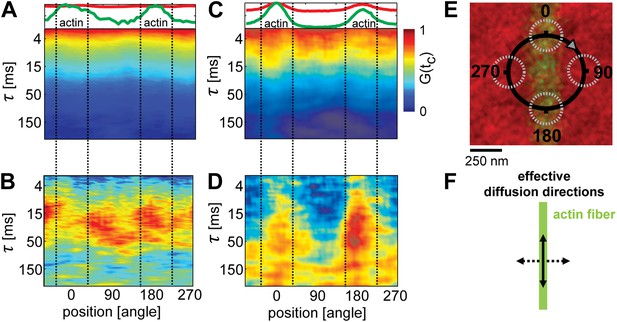
Lipid diffusion was restricted by actin-organized domains.
(A) Scanning-FCS (mobility) analysis of the Ld marker DPPE-KK114 in a single component DOPC membrane in the presence of a low density actin meshwork. The upper panel shows the intensity of the green channel (actin) and red channel (Ld marker), indicating the position of the actin fiber on the scan orbit (perpendicular lines). The lower panel depicts the auto-correlation decay for each pixel along the circular scan orbit with a diameter of d = 500 nm. The normalized auto-correlation amplitude is represented by the color. The mean transit time through the excitation spot (mobility) can be estimated by the transition from yellow to green. As expected the mobility along the scan orbit was homogenous. (B) Pair-correlation analysis of the same data as in A for opposing pixels on the scan orbit. The maxima in the pair-correlation represent the average time the probes need to move across the scan orbit. No significant directional dependence of the mobility was observed in case of simple one component membranes. (C) Same as in (A) but for ternary membranes (same composition as in Figure 1). (D) While the auto-correlation analysis seemed to be homogenous a distinct directional dependence of diffusion was revealed by the pair-correlation. In the direction along the actin fiber, a distinct correlation peak is visible with a maximum at τ ≈ 40 ms. In the perpendicular direction, the correlation amplitude is reduced, which indicates a diffusion barrier along this axis. (E) Representation of the scanning orbit over a single actin fiber bound to the membrane. The numbers represent the angles of the orbit (F). Qualitative representation summarizing the results of the pair-correlation analysis with pronounced diffusion along (drawn arrow) and restricted diffusion perpendicular to the actin fiber (dashed arrow).
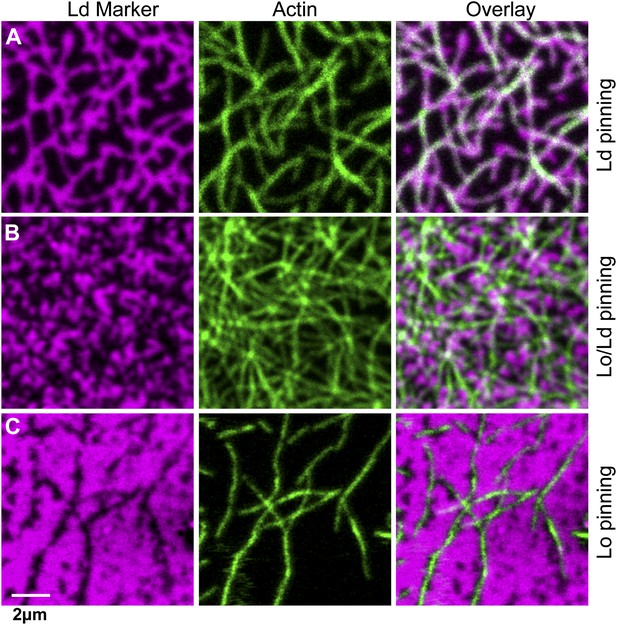
The type of pinning site used to bind actin to the membrane strongly affects the domain structure.
The Ld phase (left column, magenta) were stained with DSPE-KK114, while actin (middle column, green) was stained with phalloidin-488. The right column shows the overlay of both images. The membrane was imaged by confocal microscopy at T = 19°C, using the same lipid composition as in Figure 1. (A) Binding of actin to the Ld preferring lipid DOPE-biotin resulted in Ld domains along the actin fibers (as in Figure 1B, PCC = 0.55 ± 0.02). (B) When actin was bound to DPPE-biotin, the correlation of domains with the actin fibers was significantly reduced, but remained detectable, with a slightly positive Pearson coefficient (PCC = 0.07 ± 0.03). (C) Binding of actin to the Lo preferring lipid DSPE-PEG-biotin resulted in correlated Lo domains along the actin fibers, that is the ‘inverse’ structure of (A). In this case, the Pearson coefficient was negative (PCC = −0.37 ± 0.04). For the lipid phase partitioning values of the biotinylated lipids used in this experiment see Figure 3—figure supplement 1.
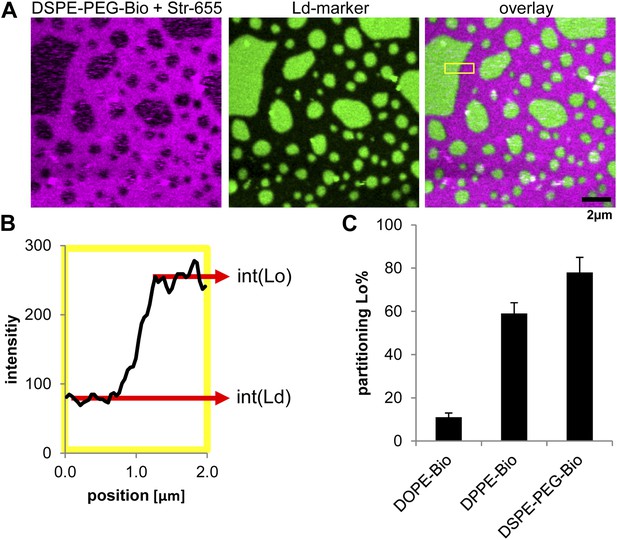
Lipid phase partitioning of biotinylated lipid–streptavidin complexes without actin.
(A) Exemplary image of a phase separated membrane (DOPC, DPPC, Cholesterol) containing 1 mol% DSPE-PEG-biotin, and the green fluorescent Ld-marker (DOPE-fluorescein; Avanti polar lipids Inc.). The biotinylated lipid was labeled by streptavidin stained with red fluorescent Atto655 (Atto-Tec GmbH, Germany). (B) The partitioning of the biotinylated lipid streptavidin complex was calculated by determining the intensity of red fluorescence in the Lo phase (int[Lo]) and in the Ld phase (int[Ld]) across a domain boundary. The scan in B corresponds to the yellow box in the overlay image. The partitioning is defined as . (C) The resulting Lo% partitioning values for the biotinylated lipids used in this study with standard variation from three independent preparations. The partitioning values serve as input parameters for our simulation model.
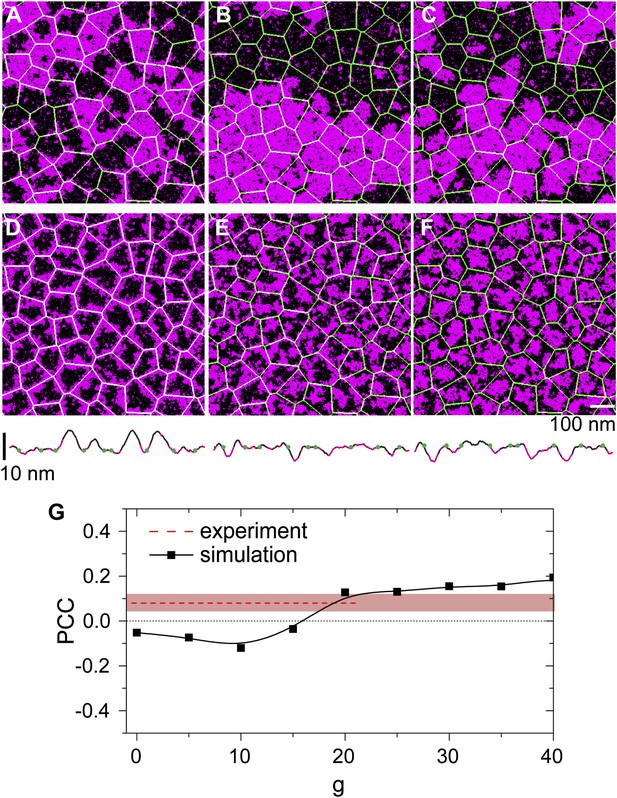
Simulation analysis of the influence of pinning and curvature coupling on lipid phase organization.
All data refer to T = 19°C. The Ld lipids are shown as magenta, the Lo lipids as black, and the actin network is shown in green as an overlay. The pinning density ρp = 0.1/nm. (A–C) Simulation snapshots obtained without coupling to curvature (g = 0) for the three species of pinning sites used in the experiments: Lo% = 11 ± 2 (A), Lo% = 59 ± 5 (B), and Lo% = 78 ± 7 (C). No significant influence of the actin network is apparent. (D–F) Same as (A–C) but in the presence of curvature coupling (g = 20). For snapshots (D) and (F), the lipid domains strongly correlate to the actin network, with Ld domains favoring actin in (D), and the inverse pattern in (F). The lower panels show height profiles of the images (D–F) scanned horizontally along the center of the image; the green dots indicate the positions of the actin fibers. (G) Pearson correlation coefficient PCC vs the curvature coupling g for the pinning species with Lo% = 59 ± 5. For weak curvature coupling, the PCC is negative indicating alignment of Lo domains along actin. By increasing the curvature coupling, the PCC becomes positive and ‘meets’ the experimentally observed value (conform Figure 4B).





