Nuclear receptor LRH-1/NR5A2 is required and targetable for liver endoplasmic reticulum stress resolution
Figures
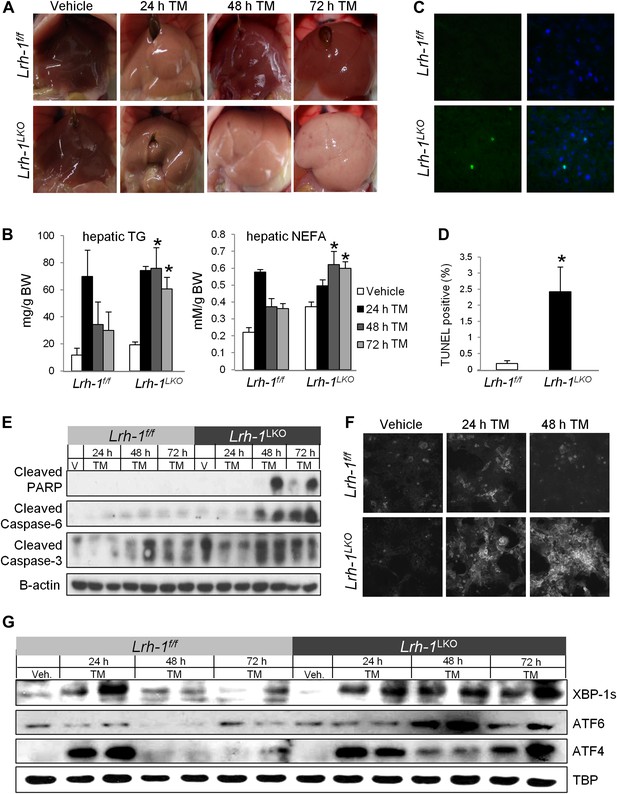
Lrh-1 is required for ER stress resolution and for protection against stress-induced lipid accumulation and cell death.
(A) Macroscopic visualization of steatosis following ER stress in Lrh-1 LKO mice. Mice were i.p. injected with 1 mg/kg tunicamycin (TM) or vehicle and livers photographed following sacrifice. Representative of 3–6 mice per group. (B) Quantification of hepatic triglycerides and non-esterified free fatty acids of control (Lrh-1f/f) and Lrh-1LKO mice (n = 3–6) injected with TM or vehicle. (C) Representative hepatic TUNEL staining for apoptosis of control and Lrh-1LKO mice injected with 1 mg/kg TM and sacrificed at 72 hr. Green fluorescence represents TUNEL-positive and blue represents DAPI-positive (merged image on right). Magnification at 40x (objective). Representative of three mice per group. (D) Quantification of TUNEL-positive cells for control and Lrh-1LKO mice treated with TM and sacrificed at 72 hr. Ratio of TUNEL-positive to DAPI-positive cells was calculated from three fields of each slide (n = 3). Significance at p<0.05. (E) Immunoblot of cleaved PARP, cleaved caspase 3, and cleaved caspase 6 in cytoplasmic fractions generated from control and Lrh-1LKO mice (n = 3–6; pooled) injected with 1 mg/kg tunicamycin (TM) or vehicle. β-actin was used as a loading control. (F) Thioflavin T fluorescence of protein aggregates in primary hepatocytes prepared from control and Lrh-1LKO mice, treated with vehicle or 0.01 µg/ml TM, and fixed in 4% PFA before staining with 500 µM Thioflavin T. Magnification at 10x (objective). Representative of three mice per group. (G) Immunoblot of nuclear spliced XBP-1, cleaved ATF6, and ATF4 for control and Lrh-1LKO mice (n = 3–6; pooled) injected with 1 mg/kg tunicamycin (TM) or vehicle. TBP was used as a loading control.
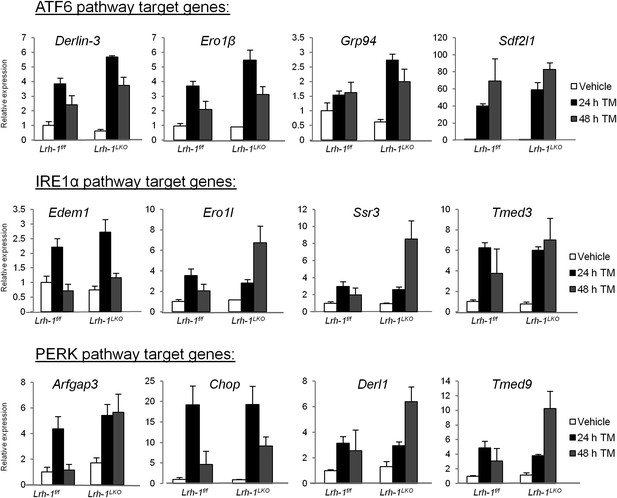
Loss of Lrh-1 does not result in loss of UPR target genes in response to stress.
Relative expression by quantitative PCR for genes dependent on each of the three UPR pathways. RNA was collected at designated timepoints for control and Lrh-1LKO mice treated with vehicle or 1 mg/kg tunicamycin (TM) (n=3-6). Data was normalized to Tbp expression.
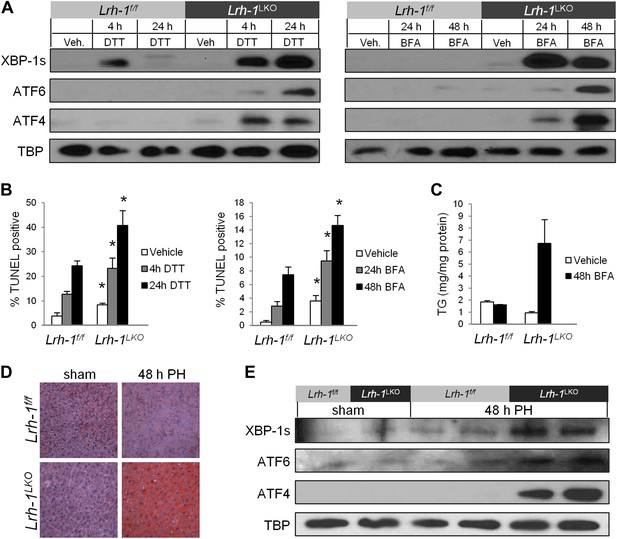
Loss of Lrh-1 sensitizes mice to ER stress resulting from chemical and physiological inducers.
(A) Immunoblot of nuclear spliced XBP-1, cleaved ATF6, and ATF4 for primary hepatocytes from control and Lrh-1LKO mice treated with 2 mM DTT or 0.05 µg/ml Brefeldin A (BFA). TBP was used as a loading control. Results representative of three independent experiments. (B) Quantification of TUNEL-positive cells for primary hepatocytes from control and Lrh-1LKO mice treated with 2 mM DTT or 0.05 µg/ml BFA. Ratio of TUNEL-positive to DAPI-positive cells was calculated from four fields of each slide (n = 3). Significance at p<0.01. (C) Quantification of triglycerides from primary hepatocytes from control and Lrh-1LKO mice (n = 3) treated with vehicle or 0.05 µg/ml BFA for 48 hr. TG was normalized to total protein by Bradford assay. (D) Partial hepatectomy (PH) was used as a non-chemical ER stress inducer. Control and Lrh-1LKO mice underwent surgical removal of 70% of liver weight or sham surgery and were sacrificed 48 hr post surgery. Oil Red O staining (200x) for neutral lipid accumulation on PH or sham surgery liver samples. Results representative of four independent samples. (E) Nuclear protein was extracted and immunoblotted for XBP-1s, ATF6, and ATF4 with TBP as a loading control for control and Lrh-1LKO mice 48 hr after sham or PH surgery. Results representative of four independent samples.
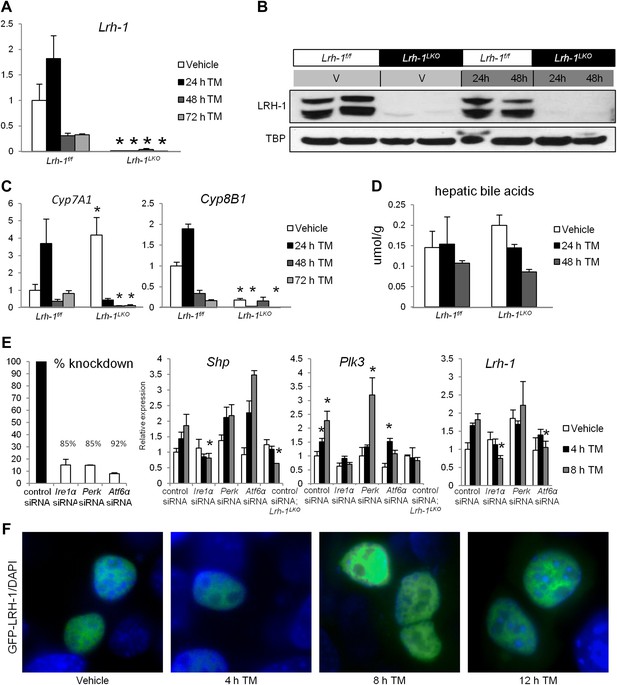
An increase in LRH-1 transcriptional activity and expression is observed following ER stress, with heightened expression dependent on UPR components.
(A) Relative expression by quantitative PCR for Lrh-1. RNA was collected at designated timepoints for control and Lrh-1LKO mice treated with vehicle or 1 mg/kg tunicamycin (TM) (n = 3–6). Data were normalized to Tbp expression. (B) Western blot analysis of nuclear LRH-1 for control and Lrh-1LKO mice injected with 1 mg/kg TM or vehicle and sacrificed at designated timepoints. TBP was used as a loading control. Results representative of 3–6 individual samples. (C) Relative expression by quantitative PCR for Cyp7A1 and Cyp8B1, which are LRH-1 target genes. RNA was collected at designated timepoints for control and Lrh-1LKO mice treated with vehicle or 1 mg/kg TM (n = 3–6). Data were normalized to Tbp expression. Significance at p<0.05 between genotypes. (D) Hepatic bile acid was extracted from tissue from control and Lrh-1LKO mice treated with vehicle or 1 mg/kg TM (n = 3–6) and sacrificed at designated timepoints. (E) Primary hepatocytes from control and Lrh-1LKO mice were transfected with siRNA against Ire1a, Perk, or Atf6, or a nonsilencing siRNA. Percent knockdown was quantified by qPCR for Ire1a, Perk, and Atf6 in control cell samples (n = 3–4) 52 hr after transfection. Relative expression for Shp, Plk3, and Lrh-1 was by qPCR for primary hepatocytes from control and Lrh-1LKO mice treated with vehicle or 5 ng/ml TM (n = 3–4). TBP was used as a loading control. Significance at p<0.05 as compared with control siRNA samples from control mice. (F) N-terminal-tagged GFP-LRH-1 fluorescence (green) in TLR-3 cells transfected with GFP-LRH-1 and treated with vehicle or 1 µg/ml TM for timepoints indicated. DNA was stained with DAPI (blue). Magnification at 100x (objective) and images cropped. Results representative of three independent experiments.
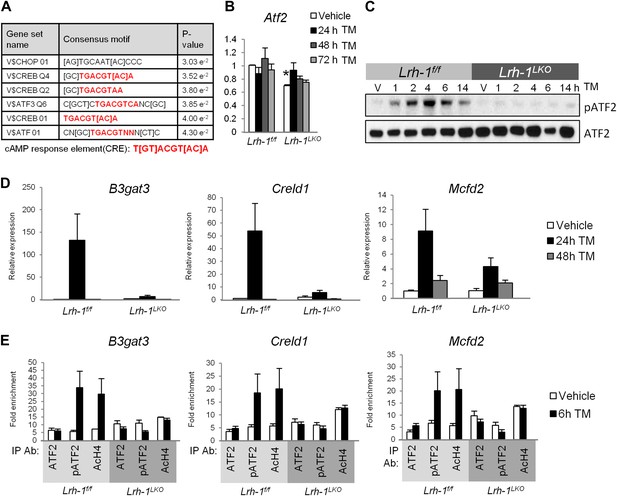
Microarray analysis suggests loss of ATF2 transcriptional ability in Lrh-1 deficient mice.
(A) Illumina Mouse Ref-8 arrays were performed for control and Lrh-1LKO mice treated with vehicle or tunicamycin (TM) for 24 hr (n = 3). Data were normalized and top 100 genes differentially induced by TM as assessed by fold change was analyzed by the Molecular Signatures Database software to identify overrepresented binding motifs. Transcription factors with binding sites significantly enriched in input gene promoters shown in chart. Motifs shown with significance at p<0.05. (B) Relative expression for Atf2. RNA was collected at designated timepoints for control and Lrh-1LKO mice treated with vehicle or 1 mg/kg TM (n = 3–6). Data were normalized to Tbp expression. Significance at p<0.01. (C) Immunoblot for total ATF2 and phospho-ATF2 (mT51/53; hT69/71), with pATF2 representing sites required to be phosphorylated for ATF2 activity. Primary hepatocytes were isolated from control and Lrh-1LKO mice and treated with 5 µg/ml TM or vehicle and protein collected at designated timepoints. Results representative of three independent experiments. (D) Relative expression for B3gat3, Creld1, and Mcfd2. RNA was collected at designated timepoints for control and Lrh-1LKO mice treated with vehicle or 1 mg/kg TM (n = 3–6). Data were normalized to Tbp expression. (E) Chromatin immunoprecipitation was performed from livers of control and Lrh-1LKO mice treated with vehicle or TM (1 mg/kg) for 6 hr (n = 4). Immunoprecipation was done with an anti-ATF2, pATF2 (69/71), or acetyl histone H4 antibody or rabbit IgG as a control. qPCR was used to determine binding by use of primers flanking binding site previously identified by ENCODE Projects.
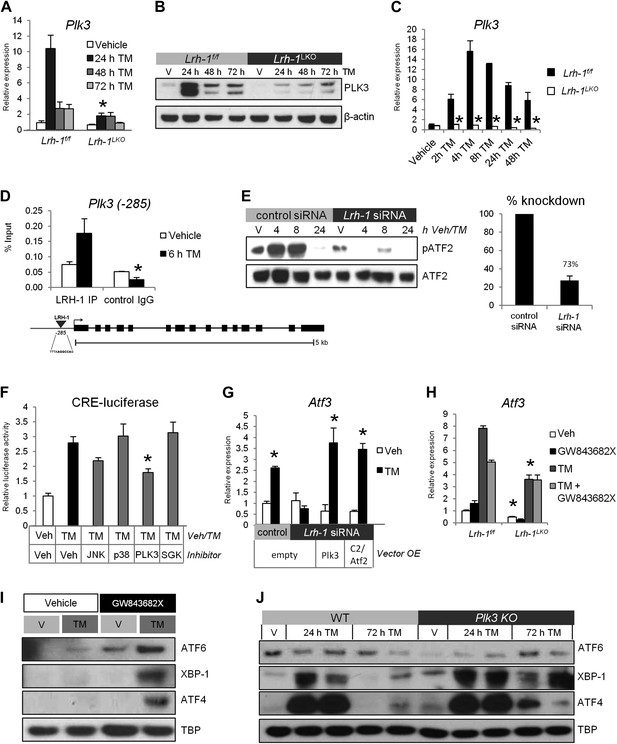
Plk3 is a direct LRH-1 target that is required for induction of ATF2 target genes and ER stress resolution.
(A) Quantitative PCR for Plk3. RNA was collected at designated timepoints for control and Lrh-1LKO mice treated with vehicle or 1 mg/kg tunicamycin (TM) (n = 3–6). Data were normalized to Tbp expression. Significance at p<0.01. (B) Immunoblot for PLK3 protein in control and Lrh-1LKO mice treated with vehicle or 1 mg/kg TM for designated timepoints. Liver tissue was boiled in Laemmli buffer to extract insoluble protein and samples were pooled (n = 2) prior to gel loading. β-actin was used as a loading control. (C) Quantitative PCR for Plk3. RNA was collected at designated timepoints for primary hepatocytes isolated from control and Lrh-1LKO mice treated with vehicle or 0.5 µg/ml TM (n = 3). Data were normalized to Tbp expression. Significance at p<0.01. (D) A LRH-1 binding site in the Plk3 promoter was identified 285 bases upstream of the TSS. Chromatin immunoprecipitation was performed from livers of control mice treated with vehicle or TM (1 mg/kg) for 6 hr (n = 3). Immunoprecipation was done with an anti-LRH-1 antibody or mouse IgG as a control. qPCR was used to determine binding by use of primers flanking binding site. Significance at p<0.05 between antibodies with error bars representing SEM. (E) TLR-3 cells were transfected with nonsilencing siRNA or siRNA against Lrh-1 and nuclear protein was prepared at designated timepoints. Samples were immunoblotted for total ATF2 and phospho-ATF2 (mT51/53; hT69/T71) following treatment with vehicle or 1 ug/ml TM. Results representative of three independent experiments. (F) Relative luciferase activity for TLR-3 cells transfected with a cAMP response element (CRE)-luciferase reporter, Atf2, and Lrh-1. 48 hr after transfection, cells were treated with vehicle or 5 µg/ml TM and the following inhibitors: 10 µM D-JNKi for JNKs, 1 µM SB202190 for p38, 10 µM GW84362X for PLK1/PKL3, or 1 µM GSK650394A for SGK. 24 hr after treatment, cells were lysed, and luciferase activity was measured and normalized to β-galactosidase activity. Significance at p<0.01 as compared with TM treated cells (n = 3). Results representative of three independent experiments. (G) Atf3 expression by qPCR in TLR-3 cells transfected with control or siRNA targeting Lrh-1 (knockdown efficiency same as 5E), along with overexpression of constitutively active Atf2 (C2/Atf2), Plk3, or an empty vector. 48 hr post transfection, cells were treated with 1 µg/ml TM for 24 hr. Data were normalized to Tbp expression. Significance at p<0.01 for TM treated vs vehicle treated samples (n = 3). Results representative of three independent experiments. (H) Atf3 expression by qPCR from primary hepatocytes from control and Lrh-1LKO mice treated with vehicle or 5 µg/ml TM and 10 µM PLK3 inhibitor GW843682X for 24 hr. Data were normalized to Tbp expression. Significance at p<0.01 between genotypes (n = cells from 2–3 mice/group). (I) Wild-type mice were i.p. injected with PLK3 inhibitor GW843682X (1 mg/kg BW) or vehicle (DMSO). Mice were also i.p. injected with TM (1 mg/kg BW) or vehicle (DMSO). 48-hr post injection, liver tissue was collected and nuclear protein was isolated to assess accumulation of UPR transcription factors spliced XBP-1, cleaved ATF6, and ATF4. TBP was used as a loading control. Results representative of results from three mice. (J) Wild-type and Plk3−/− mice were treated with TM (0.5 mg/kg) or vehicle. Nuclear UPR proteins were assessed by immunoblot at designated timepoints . TBP was used as a loading control. Results representative of four independent samples.
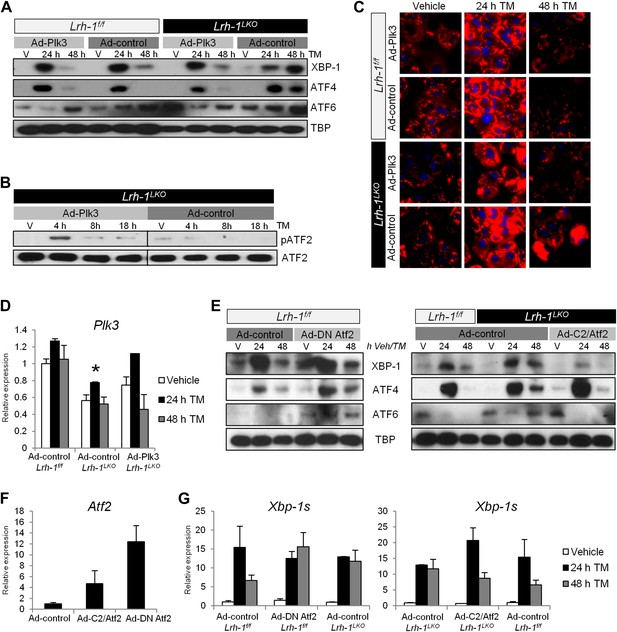
Restoration of Plk3 induction rescues ATF2 phosphorylation and ER stress resolution in Lrh-1LKO mice, and loss or gain of ATF2 transcriptional activity also alters ER stress resolution capacity.
(A) Primary hepatocytes were prepared from Lrh-1f/f and Lrh-1LKO mice and transduced with Ad-Plk3 or Ad-control at a MOI of 100. Cells were treated 36 hr later with vehicle or tunicamycin (TM) (0.01 µg/ml) and doxycycline (1 µg/ml) to induce Plk3 or LacZ control. Nuclear protein was obtained at timepoints indicated and immunoblotted for UPR transcription factors, with TBP as a loading control. Results are representative of three independent experiments. (B) Primary hepatocytes were prepared from Lrh-1LKO mice and transduced with Ad-Plk3 or Ad-control at a MOI of 100. Cells were treated 36 hr later with vehicle or TM (1 µg/ml) and doxycycline (1 µg/ml) to induce Plk3 or LacZ control. Nuclear protein was collected at indicated timepoints and immunoblotted for pATF2 (69/71) and total ATF2. Samples are from same gel and brightness/contrast was adjusted equally prior to cropping. Results are representative of three independent experiments. (C) Primary hepatocytes were prepared from Lrh-1f/f and Lrh-1LKO mice and transduced with Ad-Plk3 or Ad-control at a MOI of 100. Cells were treated 36 hr later with vehicle or TM (0.01 µg/ml) and doxycycline (1 µg/ml) to induce Plk3 or LacZ control. Cells were fixed at various timepoints and stained for lipids (red) using Lipidtox dye and counterstained with DAPI. Magnification is 40X (objective). Results are representative of three independent experiments. (D) Relative expression of Plk3 by qPCR. Primary hepatocytes from control and Lrh-1LKO mice were transduced with Ad-Plk3 or Ad-control at a MOI of 100 and treated with 1 µg/ml doxycycline and vehicle or 0.01 µg/ml TM. Data were normalized to Tbp expression. Significance at p<0.01 between genotypes (n = cells from 3–4 mice). (E) Primary hepatocytes from control mice were transduced with Ad-DN Atf2 (Atf2 T51A/T53A to serve as a dominant negative) or Ad-control at a MOI of 100 and treated with 5 ng/ml TM. Primary hepatocytes from control and Lrh-1LKO mice were transduced with Ad-C2/Atf2 (expressing a constitutively active Atf2) or Ad-control at a MOI of 100 and treated with 5 ng/ml TM. Nuclear protein was obtained at timepoints indicated and immunoblotted for UPR transcription factors, with TBP as a loading control. Results are representative of samples from three mice. (F) Atf2 expression by qPCR to quantify viral Atf2 overexpression. Primers were chosen to amplify regions identical between wildtype Atf2, C2/Atf2, and DN Atf2. Primary hepatocytes from control mice (n = 3–4) were transduced at a MOI of 100 and treated with vehicle for 24 hr prior to RNA collection. (G) Spliced Xbp-1 expression by qPCR for primary hepatocytes from control and Lrh-1LKO mice (n = 3–4) transduced with Ad-control, Ad-DN Atf2, or Ad-C2/Atf2 and treated with vehicle or 5 ng/µl TM. Data were normalized to Tbp expression.
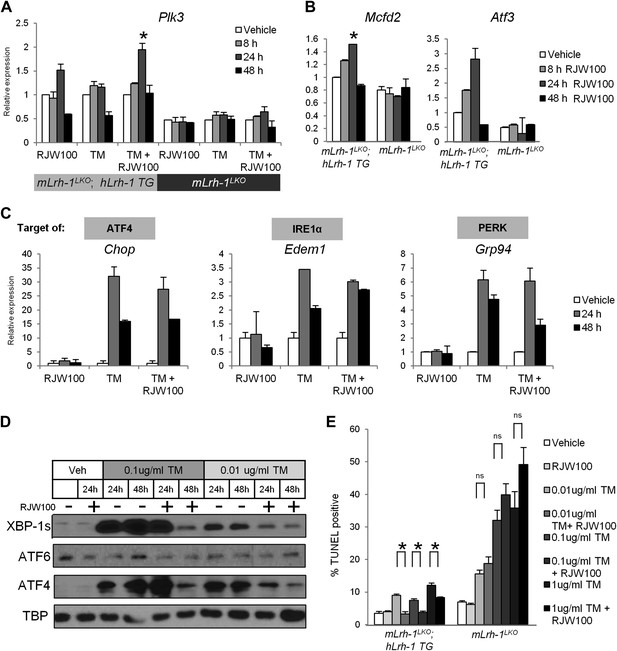
LRH-1 agonism promotes Plk3 and ATF2 target gene expression and increases resistance to ER stress independent of the UPR.
(A) Quantitative PCR for Plk3. RNA was collected at designated timepoints for primary hepatocytes isolated from hLrh-1 TG; mLrh-1 LKO and mLrh-1LKO mice treated with 10 uM RJW100 and/or 0.01 ug/ml TM (n = cells from 3 mice). Data were normalized to Tbp expression. Significance at p<0.05 as compared with vehicle treatment. (B) Quantitative PCR for Mcfd2 and Atf3. RNA was collected at designated timepoints for primary hepatocytes isolated from hLrh-1 TG; mLrh-1 LKO and mLrh-1LKO mice treated with 10 µM RJW100 (n = cells from 3 mice). Data were normalized to Tbp expression. Significance at p<0.05 as compared with vehicle treatment. (C) Primary hepatocytes were isolated from hLrh-1 TG; mLrh-1 LKO mice (n = cells from 3 mice) and treated with 10 µM RJW100 and/or 0.01 µg/ml TM. RNA was collected at designated timepoints and qPCR performed for UPR target genes. Data were normalized to Tbp expression. No significance between groups. (D) Primary hepatocytes were isolated from hLrh-1 TG; mLrh-1 LKO mice and treated with 10 µM RJW100 and/or 0.01 µg/ml or 0.1 µg/ml TM. Nuclear protein was obtained at timepoints indicated and immunoblotted for UPR transcription factors, with TBP as a loading control. Results are representative of three independent experiments. (E) Primary hepatocytes were isolated from hLrh-1 TG; mLrh-1 LKO and mLrh-1LKO mice (n = cells from three mice) and treated with 10 µM RJW100 and/or 0.01–1 µg/ml TM. Cells were fixed 48 hr post treatment and TUNEL staining was performed and quantified. Ratio of TUNEL-positive to DAPI-positive cells was calculated from four fields of each slide (n = 3). Significance at p<0.01.
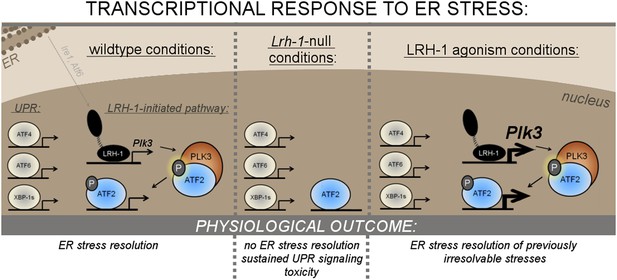
Mechanism of LRH-1’s requirement in ER stress resolution.
https://doi.org/10.7554/eLife.01694.013Tables
Genes differentially induced by TM between control and Lrh-1-null mice and their regulation by ATF2 and LRH-1
| Gene | Known target of | |
|---|---|---|
| ATF2 | LRH-1 | |
| PLK3 | ✓ | ✓ |
| GDF15 | ||
| FGF21 | * | |
| PMM1 | ||
| CRELD2 | ✓ | |
| NFIL3 | ||
| IGSF11 | ||
| GSTM2 | ||
| GOT1 | ||
| DPP9 | ✓ | ✓ |
| ST5 | ||
| CCBL1 | ✓ | ✓ |
| TMEM66 | ✓ | |
| KRTCAP2 | ✓ | ✓ |
| LRRC59 | ✓ | ✓ |
| CDK2AP2 | ✓ | ✓ |
| ST3GAL1 | ||
| TES | ✓ | |
| CCDC134 | ✓ | ✓ |
| UGT1L | ||
| ACOT2 | ✓ | |
| PLIN5 | ✓ | |
| LITAF | ✓ | |
| RPS13 | ✓ | |
| LGALS3BP | ||
| CRYM | ||
| SUPT5H | ✓ | |
| B3GAT3 | ✓ | |
| GRN | ✓ | |
| ARRDC4 | ||
| SLC30A7 | ✓ | |
| CRELD1 | ✓ | |
| NT5M | ||
| ARSG | ||
| SEP15 | ||
| MKNK1 | ✓ | |
| DDX52 | ||
| EXOSC5 | ✓ | |
| RABAC1 | ✓ | |
| RPS15 | ✓ | ✓ |
| D830014E11RIK | ||
| AVPI1 | ✓ | |
| RPS9 | ✓ | |
| CRAT | ✓ | |
| BHMT2 | ✓ | |
| B230217C12RIK | ||
| EIF3G | ✓ | ✓ |
| SLC25A28 | ✓ | |
| HR | ||
| CCL9 | ||
| ANXA4 | ✓ | ✓ |
| SMCO4 | ||
| SMOX | ||
| ARL14EP | ✓ | |
| SLC39A7 | ✓ | |
| ICA1 | ||
| ENTPD5 | ✓ | |
| PIWIL2 | ||
| ANG | ✓ | |
| MCFD2 | ✓ | ✓ |
| SOAT2 | ||
| SLC41A3 | ✓ | ✓ |
| MFGE8 | ||
| CYP4A14 | ||
| D12ERTD647E | ||
-
Microarray analysis was performed for control and Lrh-1LKO mice treated with vehicle or 1 mg/kg tunicamycin for 24 hr (n = 3). Genes were screened for those induced at least 1.5 fold by TM in control mice with significantly different induction in Lrh-1LKO mice by t-test (p<0.05). This list was filtered for those with differential expression between genotypes with TM treatment by t-test (p<0.05). Previously published genome-wide ATF2 and LRH-1 binding datasets were analyzed to identify genes with ATF2 or LRH-1 binding sites −500 to +275 bp from the TSS. Genes in our set were compared with these sets and genes that contain ATF2 or LRH-1 binding sites meeting the above criteria are marked.
-
*
No binding site for ENCODE data set but a known ATF2 direct target (Hondares et al., 2011).
Enrichment of known transcription factor binding sites in our gene set (Table 1) of differentially regulated genes by ER stress between control and Lrh-1LKO mice
| Transcription factor name: | Known phosphorylation by PLK3: | Overlap of target genes with our gene set (Table 1): | Dataset used for analysis: |
|---|---|---|---|
| ATF2 | T71 (Wang et al., 2011b) | 3.83E-08 | ATF2 binding in G12878 cells (ENCODE EH002306) |
| Lrh-1 | none | 1.40E-04 | Lrh-1 binding in mouse liver (Chong et al., 2012) |
| p53 | S20 (Xie et al., 2001) | 1.37E-04 | p53 binding in U2OS cells treated with Nutlin-3 (Menendez et al., 2013), which results in S20 phosphorylation (Valentine et al., 2011) |
| c-Jun | S63 and S73 (Wang et al., 2011a) | 2.95E-02 | c-Jun binding in CH12 cells (ENCODE EM001943), in which S63 may be constitutively phosphorylated like other B-lymphoma lines (Gururajan et al., 2005) |
| NRF2 | None | NS | NRF2 binding in lymphoid cell lines treated with sulforaphane (Chorley et al., 2012) |
-
Transcription factors known to be phosphorylated by PLK3 (ATF2, p53, and c-Jun) are summarized here, along with NRF2 to represent the oxidative stress response and LRH-1. Overlap of known transcription factor binding sites −500 to +b250 bp of the TSS for genes in Table 1 was calculated, and significance was determined using hypergeometric distribution tests.
Additional files
-
Supplementary file 1
Primer sequences.
- https://doi.org/10.7554/eLife.01694.014





