How HIV-1 Nef hijacks the AP-2 clathrin adaptor to downregulate CD4
Figures
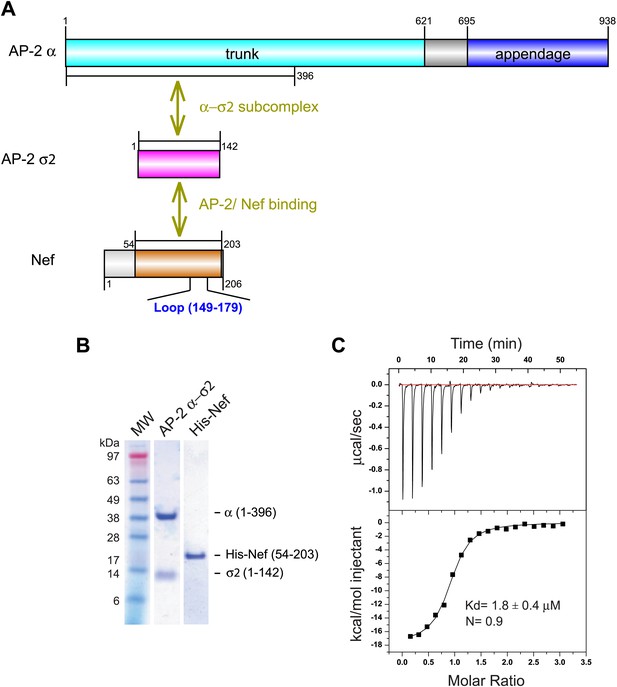
Nef binds with low micromolar affinity to the AP-2 α–σ2 hemicomplex.
(A) Schematic representation of AP-2 α–σ2 and Nef protein constructs. AP-2 α (1–396) (cyan) and full-length σ2 (magenta) were generated as a stable subcomplex and the interaction with the indicated Nef construct (54–203) (orange) was analyzed. (B) SDS gel of recombinant AP-2 α–σ2 and Nef proteins. (C) Isothermal titration calorimetry of the titration of His-tagged Nef (54–203) to the AP-2 α–σ2 hemicomplex. The upper panel shows the differential heat released when Nef (0.6 mM) was injected into AP-2 α–σ2 solution (40 μM) in 2.1 μl aliquots.
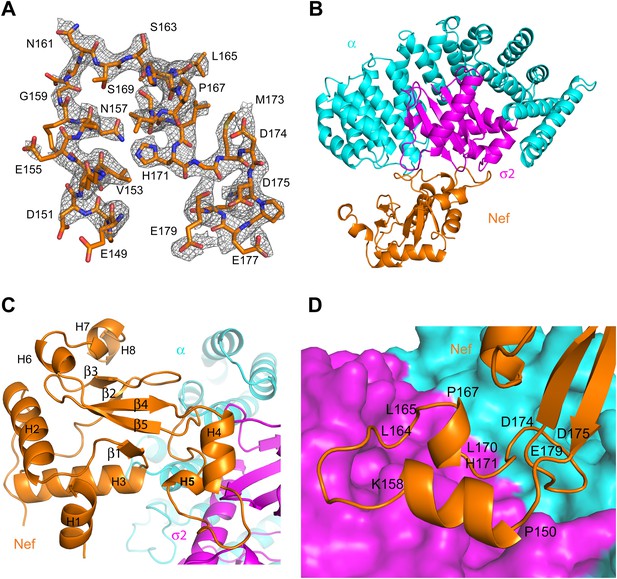
Crystal structure of the AP-2 α–σ2:Nef complex.
(A) F0-Fc omit map of Nef loop (149–179) with the final model superimposed. The map is contoured at 2.0 σ. (B) Overall ribbon representation of AP-2 α (cyan) and AP-2 σ2 subunits (magenta) in complex with Nef (orange). (C) Detailed ribbon model of Nef (orange) with the secondary structures indicated. (D) Ribbon model of the Nef central loop (149–179), which includes helix H4 (150–157), the acidic-dileucine motif (160ExxxLL165), helix H5 (167–170), and the C-terminal turn-rich segment (171–179).
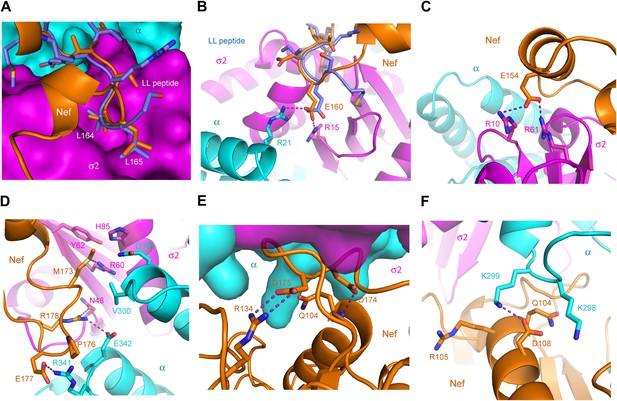
The AP-2 α–σ2:Nef interface.
(A) Stick representation of the Nef dileucine motif (Leu164 and Leu165, orange) interacting with AP-2 σ2 (magenta), compared with a bound dileucine peptide (blue, PDB id: 2JKR) (Kelly et al., 2008). (B) Nef Glu160 of the acidic-dileucine motif forms hydrogen bonds with AP-2 α R21 and σ2 R15. The hydrogen bond is listed as a purple dashed line. (C) Nef Glu154 in helix H4 (orange) forms hydrogen bonds with AP-2 σ2 R10 and R61 (magenta). (D) The C-terminal part of Nef loop (171–178) interacts with both AP-2 α and σ2 (magenta). (E) The key Nef diacidic motif Asp174 and Asp175 forms intramolecular hydrogen bonds that stabilize the loop conformation. Hydrogen bonds occur between the side chain of Asp174 and the main-chain amide NH of Gln104, and between the side-chains of Nef Asp175 and Arg134. (F) A salt bridge between Asp108 of Nef helix H3 bridges the Nef core to a basic patch on α.
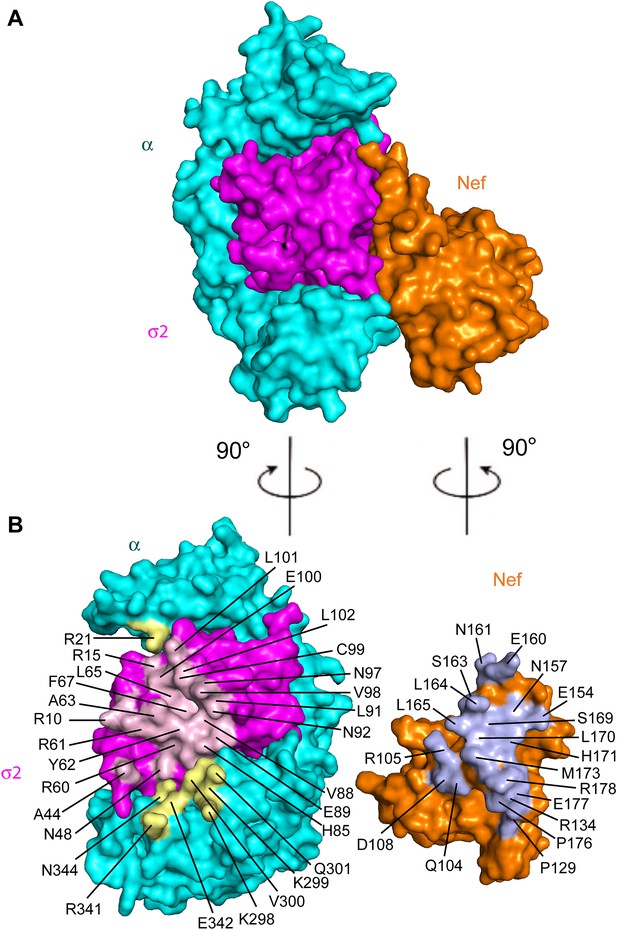
Structural mapping of mutations that interfere with binding and CD4 downregulation.
(A) The surface representation shows the contact between AP-2 α−σ2 and Nef. (B) AP-2 α−σ2 or Nef interfaces are rotated by 90° to expose the interaction surfaces directly to view. Interacting residues in AP-2 α are colored in yellow, residues in AP-2 σ2 are colored in pink, and residues in HIV-1 Nef are highlighted in light blue.
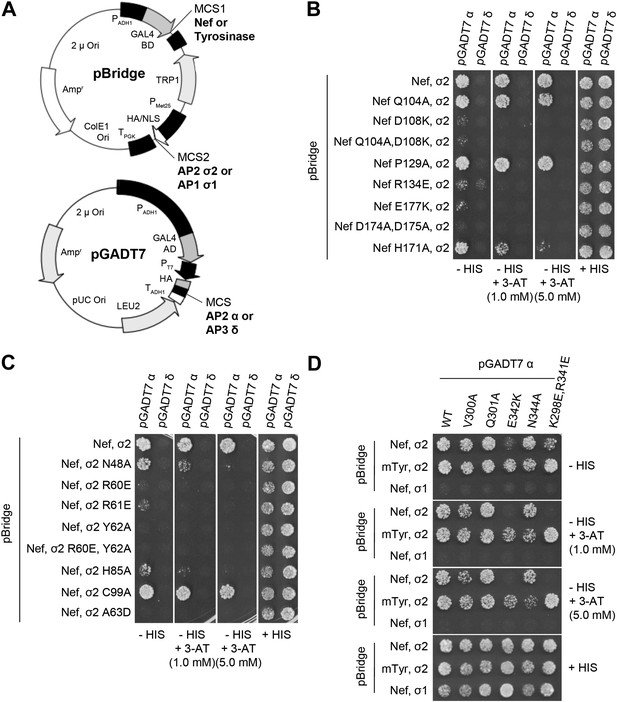
Structural interface mutants of Nef and AP-2 complexes prevent binding.
Y3H analysis of HIV-1 Nef and AP-2 α–σ2 hemicomplexes with mutations of residues revealed in the crystal structure. (A) Diagram of the plasmids used in Y3H analysis. NL4-3 Nef or mouse Tyrosinase cytosolic tail was cloned into MCS1 of pBridge and expressed as a GAL4BD fusion protein. AP-2 σ2 or AP-1 σ1 was cloned into MCS2 of pBridge and expressed without Met. AP-2 α or AP-3 δ was cloned into MCS of pGADT7 and expressed as a GAL4AD fusion protein. (B–D). The indicated combinations of double transformants were plated in media lacking Leu, Trp, Met and His (−HIS),−HIS with 3-AT (1 mM or 5 mM) or Leu, Trp and Met (+HIS). mTyr, mouse Tyrosinase cytosolic domain.
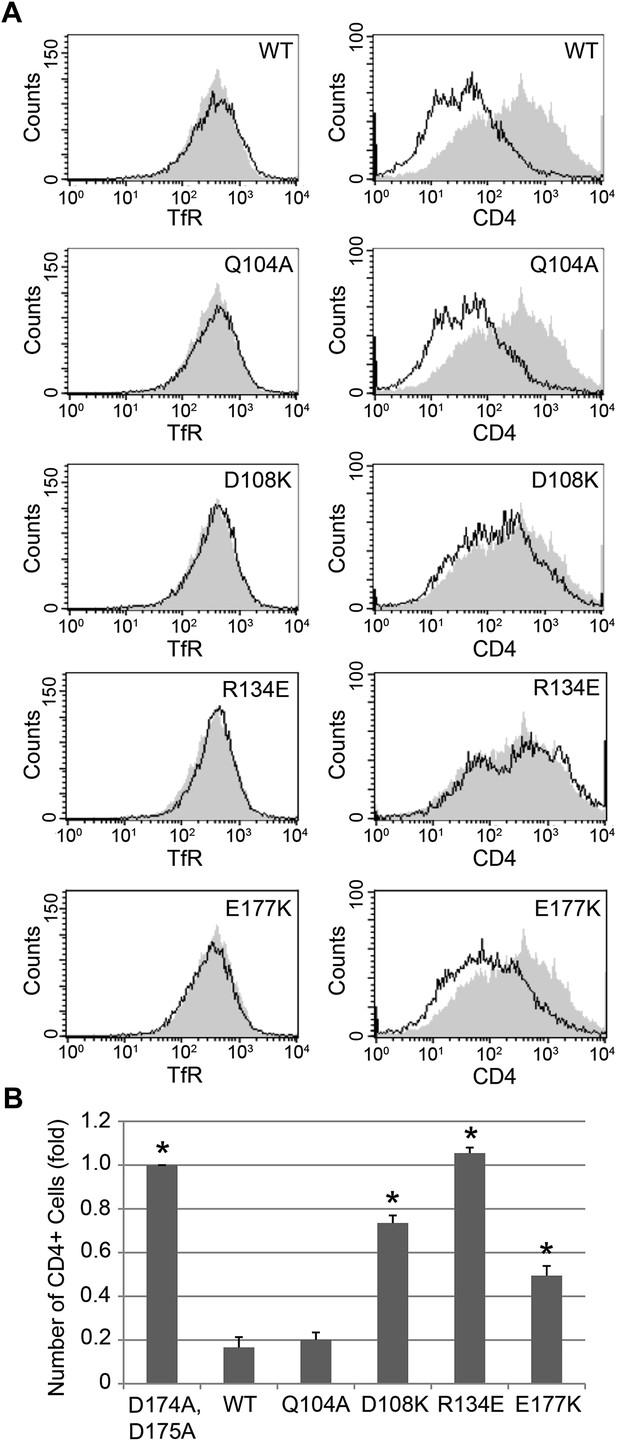
Nef interface mutants do not downregulate CD4.
Nef Asp108, Arg134, and Glu177 are required for the Nef-induced CD4 downregulation. (A) HeLa cells were cotransfected with pCMV-CD4 and pIRES-eGFP-Nef wild-type or mutant plasmids for 24 hr. The cells were then stained with APC-conjugated anti-CD4 antibody and PE-conjugated anti-Transferrin receptor (TfR) antibody. GFP was used as an indicator for transfected cells. The D174A, D175A mutant Nef was used as a negative control (Shaded curves in all plots). Data shown are representative of three independent experiments. (B) The graph shows the relative number of CD4 positive cells from Figure 6A (mean ± SD; N = 3; asterisks: p<0.001 compared with wild-type Nef).
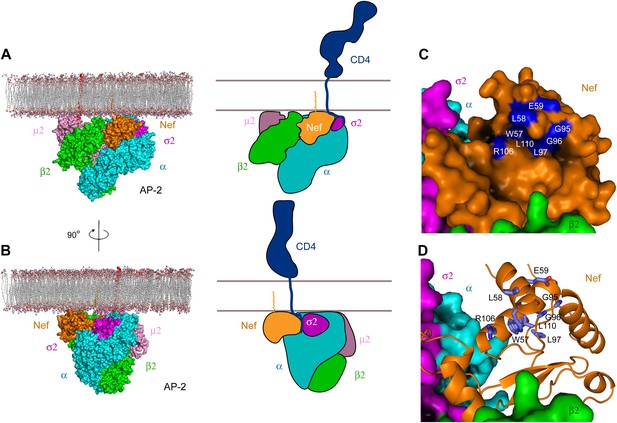
Docking of the unlocked AP-2:Nef complex to the membrane.
(A) The unlocked conformation of AP-2 core bound to myristoylated Nef (orange). The AP-2 α–σ2:Nef structure was first aligned with the open conformation of AP-2 core structure (2XA7), and then docked on the membrane. The second view (B) is shown by rotating the first by 90°. Schematics are shown to the right of (A) and (B). (C) The surface of the AP-2:Nef complex as viewed from the membrane. Nef residues that are mapped by CD4 binding (Grzesiek et al., 1996b) are colored in blue. (D) Stick representation of Nef residues (blue) that interact with the CD4 cytosolic tail.
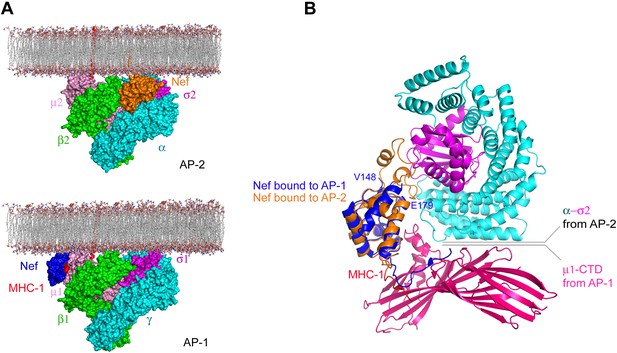
Nef uses different surfaces to bind to different regions of AP-1 and AP-2.
(A) Nef binds to different subunits of AP-2 (top) and AP-1 (bottom), but docks onto the membrane in both cases. The MHC-I cytoplasmic domain (CD):Nef complex binds to the μ1 C-terminal domain (CTD) of the AP-1 core. One copy of AP-1 μ1-CTD:MHC-I-CD:Nef complex (Jia et al., 2012) (pdb: 4EN2) was aligned with the open conformation of AP-1 core structure (Ren et al., 2013) (pdb: 4HMY), and then the AP-1 complex was docked onto the membrane in the same orientation as shown for AP-2 in Figure 7 and in the top panel. (B) Structural superposition of Nef (blue) as bound to the μ1 subunit of AP-1 upon Nef (orange) bound to the α–σ2 hemicomplex of AP-2 in this study.
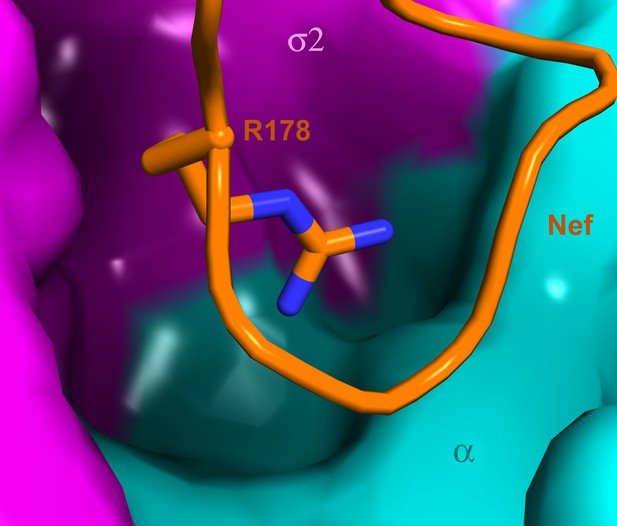
A highly concave pocket specific for the Nef interaction.
Nef is shown in a stick model and the highly concave AP-2 surface is shown in the vicinity of Nef Arg178. This region has no known interactions with physiological cargoes.
Tables
Statistics of crystallographic data collection and refinement
| Construct | AP-2: σ2 (1–143), α (1–396); Nef (54–203) |
| Data collection | |
| X-ray source | APS 22-ID |
| Wavelength (Å) | 1.0000 |
| Space group | P212121 |
| Cell dimensions | a = 109.56 Å, b = 168.03 Å, c = 200.20 Å, α = β = γ = 90° |
| Resolution (Å) (last shell) | 50.00–2.90 (3.00–2.90) |
| Unique reflections | 80,188 |
| Rsym* (%) | 18.4 (53.9) |
| I/σ | 7.4 (1.9) |
| Completeness | 95.8 (80.2) |
| Redundancy | 5.2 (3.2) |
| Refinement | |
| Rwork/Rfree (%) | 21.9/26.7 |
| Average B values (Å2) | 39.5 |
| Number of protein atoms | 21,588 |
| R.m.s. bond length deviation (Å) | 0.015 |
| R.m.s. bond angle deviation (°) | 1.16 |
| Ramachandran Plot (%) | |
| Favored | 98.4 |
| allowed | 1.2 |
| outlier | 0.4 |
-
*
Rsym = ΣhΣi|Ii(h)−<I>|/ΣhΣiIi(h), where I is the observed intensity and <I> is the average intensity of multiple observations of symmetry-related reflections.
Functional importance of Nef-AP-2 interacting residues
| Nef residue | References | Interacting residues | Interactions | References |
|---|---|---|---|---|
| Q104 | This study | α K298*, K299 | Hydrogen bond | (Chaudhuri et al., 2009) |
| D108* | This study | α K298*, K299 | Salt bridge | (Chaudhuri et al., 2009) |
| P129 | This study | α R341* | Van der Waals | (Chaudhuri et al., 2009) |
| R134* | This study | Nef D175* | Nef core-to-loop internal salt bridge | This study; (Mattera et al., 2011) |
| E154 | (Lindwasser et al., 2008) | σ2 R61, R10 | Salt bridge | This study |
| N157 | (Lindwasser et al., 2008) | σ2 A63 | Van der Waals | (Mattera et al., 2011) |
| E160 | (Lindwasser et al., 2008) | σ2 R15, α R21 | Salt bridge | (Mattera et al., 2011) |
| N161 | (Lindwasser et al., 2008) | σ2 C99, L101 | Van der Waals | This study; (Mattera et al., 2011) |
| S163 | (Lindwasser et al., 2008) | σ2 N97 | Weak hydrogen bonds | |
| L164* | (Janvier et al., 2003; Lindwasser et al., 2008) | σ2 Y62, A63, L65, F67, V88, L91, V98, L103 | Hydrophobic | This study; (Mattera et al., 2011) |
| L165* | (Janvier et al., 2003; Lindwasser et al., 2008) | σ2 Y62, H85, V88, E89, N92 | Hydrophobic | This study; (Mattera et al., 2011) |
| S169 | (Lindwasser et al., 2008) | σ2 A63 | Van der Waals | (Mattera et al., 2011) |
| L170* | (Lindwasser et al., 2008; Jin et al., 2013) | σ2 Y62 | Hydrophobic | This study |
| H171 | This study; (Lindwasser et al., 2008) | σ2 A63 | Hydrogen bond to main chain carbonyl | This study; (Mattera et al., 2011) |
| M173 | (Lindwasser et al., 2008) | α Q301, V300; σ2 R60, Y62, H85 | Hydrophobic and nitrogen-sulfur hydrogen bond | This study |
| D174* | (Lindwasser et al., 2008) | Nef Q104 | Nef core-to-loop internal hydrogen bond to main chain amide | This study |
| D175* | (Lindwasser et al., 2008) | Nef R134* | Nef core-to-loop internal salt bridge | This study; (Chaudhuri et al., 2009) |
| P176 | (Lindwasser et al., 2008) | α R341*, E342 | Van der Waals | This study; (Chaudhuri et al., 2009) |
| E177* | This study; (Lindwasser et al., 2008) | α R341* | Salt bridge | This study; (Chaudhuri et al., 2009) |
| R178 | (Lindwasser et al., 2008) | α E342, σ2 N48 | Strong hydrogen bond, weak salt bridge | This study |
-
Bold: mutation inhibits binding. Italics: mutation has no effect on binding. Plain text: not tested. Only residues tested as single amino acid substitutions are included.
-
*
Mutation inhibits CD4 downregulation activity. All residues tested as single amino acid substitutions except for K298/R341, which were tested together.





