ERK8 is a negative regulator of O-GalNAc glycosylation and cell migration
Figures
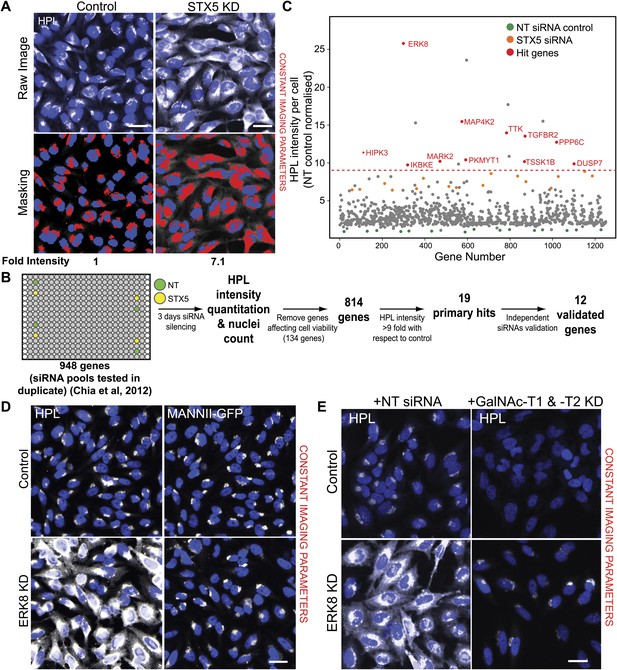
RNAi screening reveals 12 negative regulators of Tn expression.
(A) Helix pomatia lectin (HPL) staining was analysed using the ‘Transfluor HT’ module of MetaXpress software (Molecular Devices). A mask was generated for both HPL and nuclei (Hoechst) staining to classify the region of measurement (lower panels). Scale bar: 30 µm. (B) Schematic overview of the screening process. Images from the RNAi screen in Chia et al. (2012) were quantified for HPL intensities. Non-targeting (NT) siRNA and Syntaxin 5 (STX5) siRNA were used as negative and positive controls, resepectively. (C) Fold-change of HPL intensities normalised to NT siRNA treatment (green dots) and STX5 (orange dots). Primary hits were selected based on a threshold of a nine-fold increase (red dashed line) and the final validated genes are labelled in red (Hit genes). (D) Images from the screen of HPL staining in HeLa cells depleted of ERK8. MannII-GFP labels the Golgi apparatus. Scale bar: 30 μm. (E) HPL staining in cells knockdown of ERK8 with a control siRNA or GalNAc-T1 and -T2 siRNA. Scale bar: 30 μm.
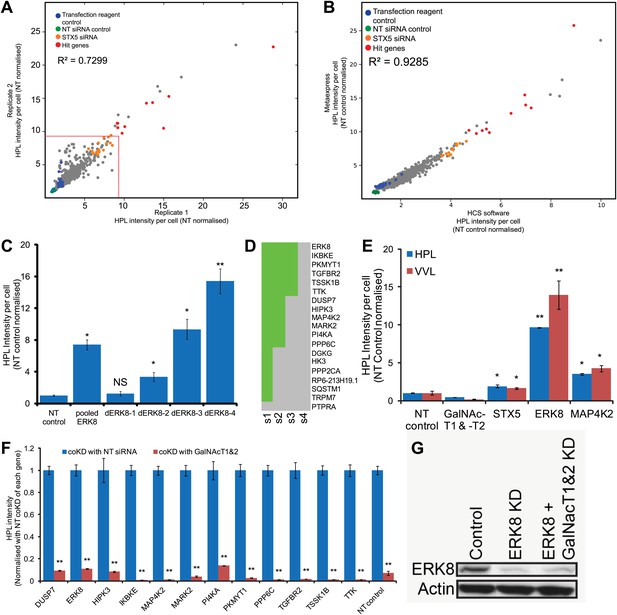
Helix Pomatia Lectin (HPL) stains reliably and specifically for Tn antigen.
(A) Comparison of the HPL intensities between the two screen replicates. (B) Comparison of HPL intensities generated from different analysis algorithms from the HCSU (high content screening unit) application (Chia et al., 2012) and MetaXpress transfluor HT (Molecular Devices). (C) Quantification of the Tn levels of ERK8 depletion with pooled and deconvoluted siRNAs (dERK8-1 to 4). (D) Hit validation using deconvoluted siRNA pools. HPL intensities were quantified in HeLa cells treated with each of the four individual duplex siRNAs from the pool for the 19 primary hits. A gene was validated if at least two unique siRNAs reproduced at least a 4.5-fold increase in HPL intensities. (E) Comparison of HPL and Vicia Villosa Lectin (VVL) staining intensities in ERK8 and MAP4K2 depletion. (F) Quantification of Tn levels upon co-knockdown (coKD) of each of the 12 validated Tn regulators with GalNAc-T1 and -T2. (G) SDS-PAGE analysis of ERK8 expression levels in single and co-knockdown with GalNAc-T1 and -T2. Values on graphs indicate mean ± SEM. **p<0.0001, *p<0.05 by two-tailed unpaired t test, relative to the non-targeting (NT) siRNA-treated cells. NS, not significant.
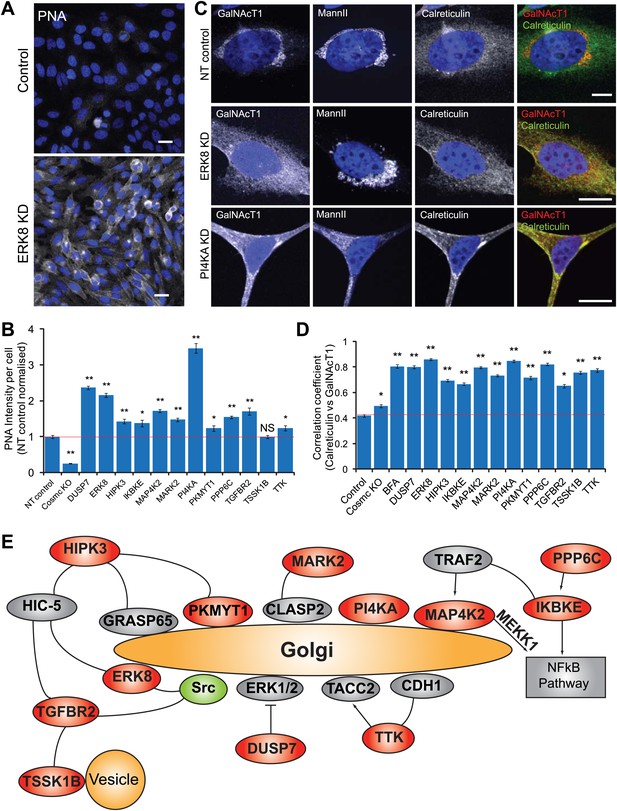
Tn regulators control Tn expression through GalNAc-T subcellular localisation.
(A) Peanut Agglutinin (PNA) lectin staining in ERK8-depleted HeLa cells. Scale bar: 30 μm. (B) PNA lectin staining quantification after depletion of the 12 Tn regulators, using COSMC knockout HeLa cells as a positive control. (C) Co-staining for endogenous GalNAc-T1 and Golgi (MannII-GFP) and ER (Calreticulin) markers. Scale bar: 10 μm. (D) Co-localisation of the GalNAc-T1 and Calreticulin measured using Pearson’s correlation coefficient of the staining intensities of the two markers. Cells were analysed using MetaXpress Translocation-Enhanced analysis module. Values on graphs indicate the mean ± SEM. **p<0.0001, *p<0.05 by two-tailed unpaired t test, relative to NT siRNA-treated cells. (E) A potential regulatory network of signalling proteins regulating GalNAc-T localisation.
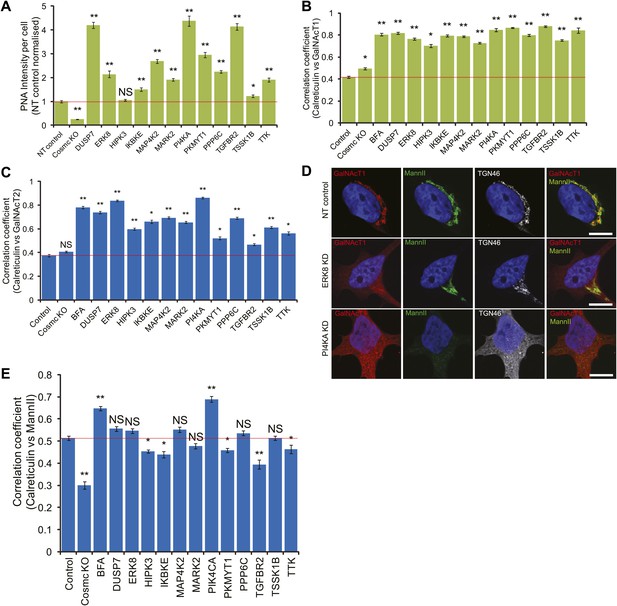
Tn regulators control GalNAc-T1 and -T2 localisation.
(A) Peanut Agglutinin (PNA) lectin staining quantification after depletion of the 12 Tn regulators with a single siRNA from the pool used in the screen and in COSMC knockout cells. (B) Pearson's correlation coefficient between GalNAc-T1 and Calreticulin staining in cells knocked down with a single siRNA from the pool. (C) Pearson's correlation coefficient between GalNAc-T2 and Calreticulin staining in the depletion of each of the 12 Tn regulators. (D) Co-staining for endogenous GalNAc-T1, medial Golgi (MannII-GFP) and trans Golgi (TGN46) markers in NT, ERK8-KD and PI4KA-KD cells. Scale bar: 10 μm. (E) Pearson's correlation coefficient between MannII-GFP and Calreticulin staining in the depletion of each of the 12 Tn regulators. Values on graphs indicate mean ± SEM. **p<0.0001, *p<0.05 by two-tailed unpaired t test, relative to non-targeting (NT) siRNA-treated cells. NS, not significant.
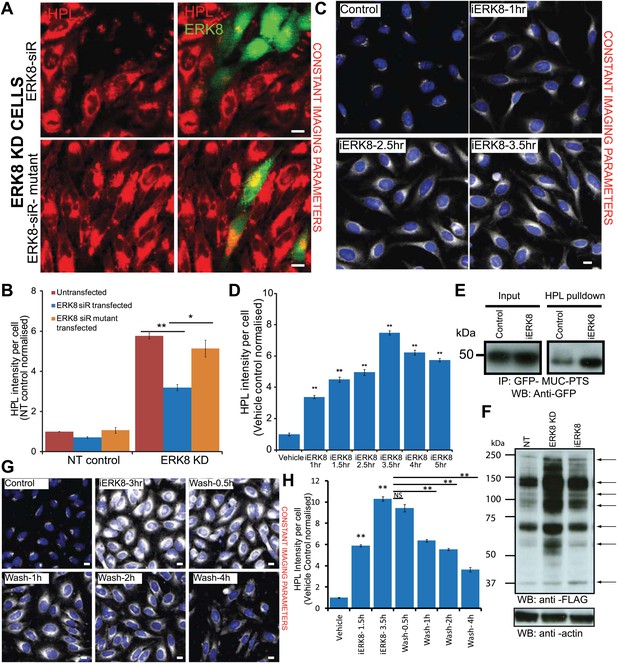
ERK8 regulates ER-localised O-glycosylation initiation.
(A) Protein replacement by expression of siRNA-resistant wild-type ERK8 or the kinase inactive mutant in ERK8-depleted HeLa cells. Cells were stained for Helix pomatia lectin (HPL) and ERK8. Scale bar: 30 µm. (B) Tn levels of non-targeting (NT) siRNA-treated and ERK8-depleted cells that were untransfected (red bars) or transfected with wild-type ERK8 (blue bars) or kinase-inactive mutant ERK8 (orange bars). (C) Treatment with 5 µM ERK8 inhibitor Ro-31-8220 (iERK8) over time and staining for Tn expression with HPL in HeLa cells. (D) Quantification of Tn expression after 5 µM iERK8 treatment. (E) SDS-PAGE analysis of ER-specific glycosylation reporter (Muc-PTS) expressed in HEK293T cells treated with vehicle or with 5 µM iERK8 for 3.5 hr. Muc-PTS was immunoprecipitated using HPL-conjugated agarose. (F) SDS-PAGE analysis of untreated, ERK8-depleted and inhibitor-treated cell lysates metabolically labelled using GalNAz-FLAG. Arrows point to bands with changed intensities (G) Tn staining after 5 µM iERK8 treatment for 3 hr followed by chase over time. Scale bar: 30 µm. (H) Quantification of Tn expression levels upon iERK8 treatment and washout. Values on graphs indicate the mean ± SEM. **p<0.0001, *p<0.05 by two-tailed unpaired t test, relative to untransfected or ERK8 wild-type transfected cells in (B) and vehicle treated cells in (D) and (H).
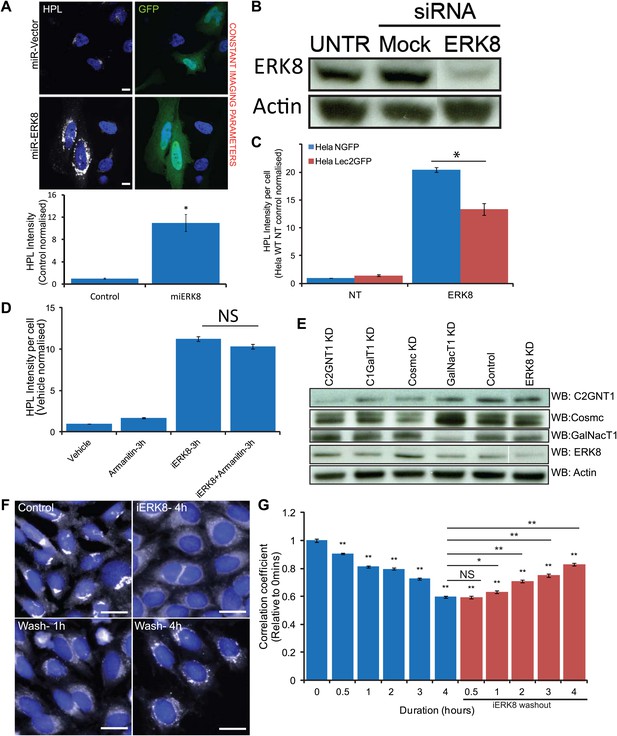
ERK8 regulates Tn expression through GalNAc-T relocation and not related to transcriptional or translational events.
(A) Tn staining with Helix Pomatia Lectin (HPL) and quantification of cells expressing mirRNA (Invitrogen) for ERK8. Scale bar: 10 μm. (B) SDS-PAGE analysis of ERK8 expression levels in cells treated with single siRNA dERK8-4. (C) Quantification of Tn levels of ERK8 depletion in ER-localised Lec2-GFP cells and GFP-expressing cells. (D) Quantification of Tn levels in cells treated with 10 µg/ml transcription inhibitor α-amanitin and co-treated with 5 µM Ro-31-8220 (iERK8). (E) SDS-PAGE analysis of protein expression levels of O-glycosylation-initiating enzymes and molecular chaperones in ERK8-depleted cells. (F) GalNAc-T1 staining of cells treated and washout (‘Wash’) of 5 µM iERK8 at the indicated times. Scale bar: 30 µm. (G) Quantification of the Pearson's correlation coefficient of GalNAc-T1 and Golgi marker MannII-GFP with iERK8 treatment (blue bars) and compound washout (red bars) after a 4-hr treatment for the indicated wash times. Values on graphs indicate the mean ± SEM. **p<0.0001, *p<0.05 by two-tailed unpaired t test, relative non-targeting (NT) control (A and C) or vehicle-treated (Control) cells (D and G). NS, not significant.
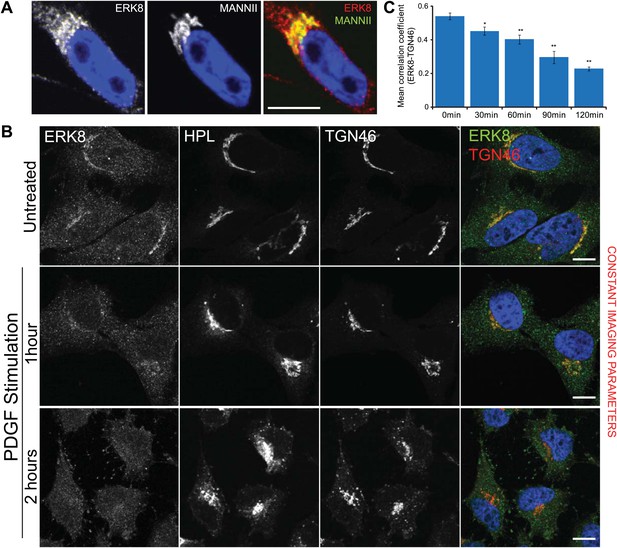
ERK8 is dynamically localised at the Golgi.
(A) High magnification of HeLa MannII-GFP expressing cells stained for endogenous ERK8 following cytosol extraction. (B) Cytosol-depleted cells treated with platelet-derived growth factor (PDGF; 50 ng/ml) for the indicated times and stained for ERK8, Tn (Helix pomatia lectin, HPL) and the Golgi marker, TGN46. Scale bar: 10 μm. (C) Pearson’s correlation coefficient between ERK8 and Golgi marker TGN46 in cells treated with PDGF for the indicated times. Values on graphs indicate the mean ± SEM. **p<0.0001, *p<0.05 by two-tailed unpaired t test, relative to vehicle treated cells.
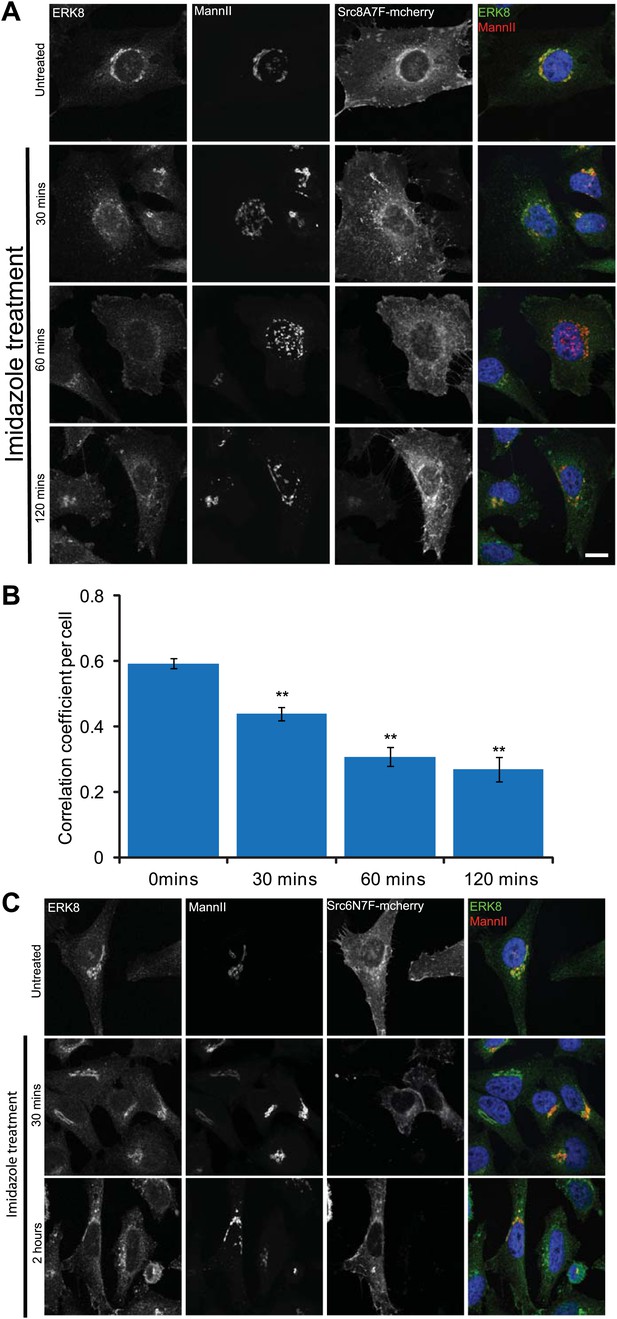
ERK8 is dynamically localised when SRC is increasingly activated at the Golgi.
(A) MannII-GFP- and Src-8A7F-mcherry-expressing cells were treated with imidazole for the indicated times, cytosol-depleted and stained for ERK8. Scale bar: 10 μm. (B) Pearson's correlation coefficient between ERK8 and Golgi marker MannII in cells treated with imidazole for the indicated times. Values on graphs indicate the mean ± SEM. **p<0.0001, *p<0.05 by two-tailed unpaired t test, relative to vehicle treated cells. (C) Catalytically defective mutant Src-6N7F-mcherry-expressing cells were treated with imidazole for indicated times, cytosol-depleted and stained for ERK8 and the Golgi marker MannII-GFP. Scale bar: 10 μm.
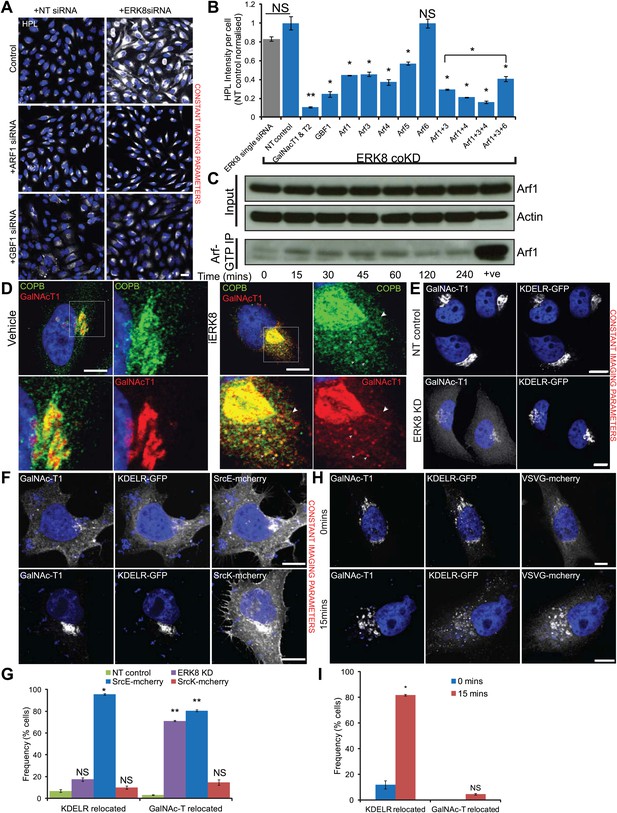
ERK8 regulates COPI-dependent GalNAc-T traffic.
(A) Co-knockdown of ERK8 with Arf1 or GTP exchange factor, GBF1, and staining with Helix pomatia lectin (HPL). NT, non-targeting. Scale bar: 30 μm. (B) Quantification of Tn levels upon ERK8 co-knockdown with Arf proteins and GBF1. Grey bar indicates knockdown of ERK8 only. Blue bars indicate co-knockdowns. (C) SDS-PAGE analysis of total Arf and Arf1-GTP in cells treated with 5 µM Ro-31-8220 (iERK8). (D) Co-staining of Beta-COP (COPB) and GalNAc-T1 in cells treated with 5 µM iERK8 for 15 min. Transient tubular structures emanating from the Golgi appear stained for GalNAc-T1 and beta-COP (arrowhead, second panel). Scale bar: 10 μm. (E) Effect of ERK8 depletion on GalNAc-T1 and KDEL receptor (KDEL-R) subcellular location. (F) Effect of expression of active SRC (SrcE-mcherry, containing the E378G mutation) or inactive SRC (SrcK-mcherry, containing the K295M mutation) on both proteins. Scale bar: 10 μm. (G) Visual scoring of KDEL-R and GalNAc-T1 redistribution from the Golgi in cells subjected to various treatment conditions. Cells were counted in each condition from three independent experiments (NT control: 83 cells; ERK8 KD: 86; SrcE-mcherry: 42; SrcK-mcherry: 32). (H) Temperature-sensitive vesicular stomatitis virus G glycoprotein (VSVG-mcherry) traffic to the Golgi induced by shift from restrictive to permissive temperature for 15 min in KDEL-R expressing cells stained for GalNac-T1. Scale bar: 10 μm. (I) Visual scoring of KDEL-R and GalNAc-T1 relocation in VSVG expressing cells at 0 and 15 min after temperature shift. Cells were counted in each condition from three independent experiments (0 min, 44 cells; 15 min, 63 cells). Values on graphs indicate the mean ± SEM. **p<0.0001, *p<0.05 by two-tailed unpaired t test, relative to NT siRNA-treated (B and G) cells and cells at 0-min timepoint (I).
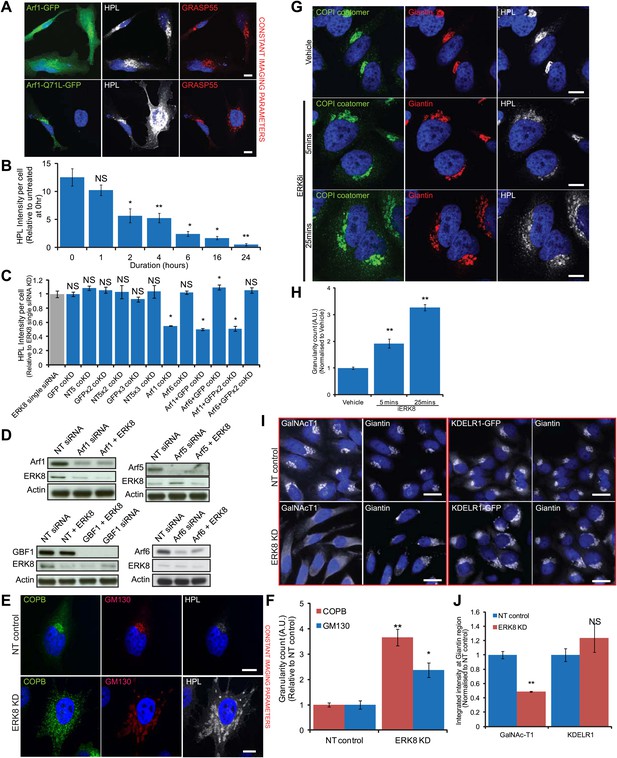
ERK8 regulated GalNAc-T traffic depends on the activity of COPI regulators.
(A) Tn expression levels, as determined by Helix Pomatia Lectin (HPL) staining in cells expressing dominant-negative mutant Arf1 (Q71L) compared to wild-type Arf1. GRASP55 labels the Golgi apparatus. Scale bar: 10 μm. (B) Quantification of Tn expression in ERK8-depleted cells upon treatment with 50 nM GBF1 inhibitor, Golgicide, for the indicated times. (C) Quantification of Tn expression in cells following co-knockdown with ERK8 and GFP or NT5 siRNA in amounts to equivalent to double (GFP coKD), triple (GFPx2 coKD) and quadruple (GFPx3 coKD) siRNA transfection configurations. (D) SDS-PAGE analysis of protein levels of Arfs and GBF1 in single and double knockdown configurations. (E) Beta-COP (COPB) staining in control and ERK8-depleted cells. Scale bar: 10 μm. (F) Quantification of the relative numbers of COPI transport carriers (COPB) and Golgi fragments (GM130) using the granularity measurement module in MetaXpress software. At least 30 cells (non-targeting (NT) control, 37 cells; ERK8-KD, 34) were quantified for each treatment. (G) Staining of native COPI coatomer in cells treated with 5 μM Ro-31-8220 (iERK8) over the indicated times. (H) Quantification of the relative numbers of COPI transport carriers using the granularity measurement module in MetaXpress software. Twenty-five or more cells (Vehicle, 41 cells; 5 min iERK8 treatment, 25; 25 min iERK8 treatment, 57) were quantified for each treatment. (I) Staining of GalNAc-T1 (left panel) and KDEL receptor (KDELR1) localisation (right panel) in HeLa KDEL-R1-GFP stable cell line depleted of ERK8. (J) Quantification of total intensities of GalNAc-T1 and KDEL-R at the Golgi region demarcated by Giantin (Golgi protein) staining. Values on graphs indicate the mean ± SEM. **p<0.0001, *p<0.05 by two-tailed unpaired t test, relative to vehicle treated (B and H), single ERK8 knockdown (C) and NT siRNA-treated (F and J) cells. NS, not significant.
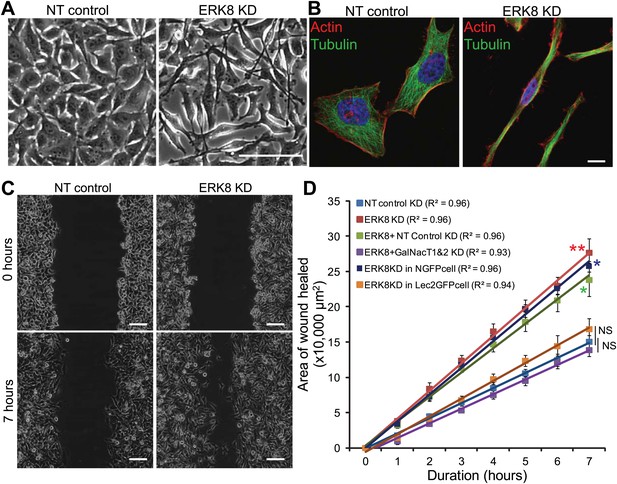
ERK8 regulates cell migration through ER O-glycosylation.
(A) Phase contrast images and (B) actin and tubulin staining of non-targeting (NT) siRNA-treated and ERK8-depleted cells. Scale bars: 100 μm in (A) and 10 μm in (B). (C) Migration assay using scratch wound of cellular monolayer in NT siRNA-treated and ERK8-depleted cells. Scale bar: 100 μm. (D) Rate of wound closure (area) measured over 7 hr (n = 4 experiments for each condition). Values on graphs indicate mean ± SEM. **p<0.001, *p<0.05 by two-tailed unpaired t test. Red asterisks indicate t test between NT siRNA-treated and ERK8-depleted cells. Green asterisk indicates t-test between cells co-knockdown with ERK8 and GalNAc-T1 and -T2 (ERK8+GalNAc-T1 & -T2 KD) and cells co-knockdown with ERK8 and NT siRNA (ERK8+NT control KD). Blue asterisk indicates t test between ERK8 knockdown in NGFP-expressing and ER-localised GalNAc-T inhibitor Lec2GFP cells. NS, not significant (black vertical lines).
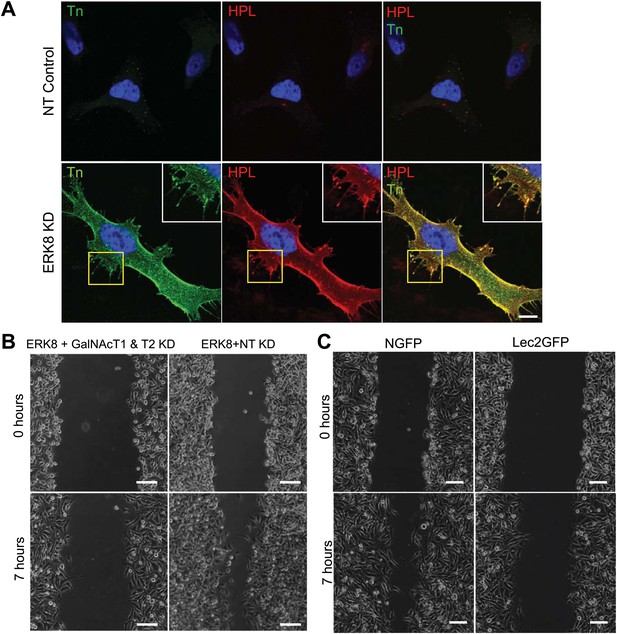
ERK8 inhibits cell motility by controlling Tn expression on cell-surface O-glycoproteins.
(A) Cell-surface staining using a specific Tn antibody (green) and Helix Pomatia Lectin (HPL) (red) on non-permeabilised ERK8-depleted cells. Scale bar: 10 μm. (B) Scratch wound assay of cellular monolayer of cells following co-knockdown with ERK8 and GalNAc-T1 and -T2 (ERK8+GalNacT1 & T2) siRNA or co-knockdown with ERK8 and non-targeting (NT) siRNA (ERK8+NT) on fibronectin-coated plates. Scale bar: 100 μm. (C) Scratch wound assay of ERK8-depleted NGFP- and Lec2GFP-expressing cells. Scale bar: 100 μm.
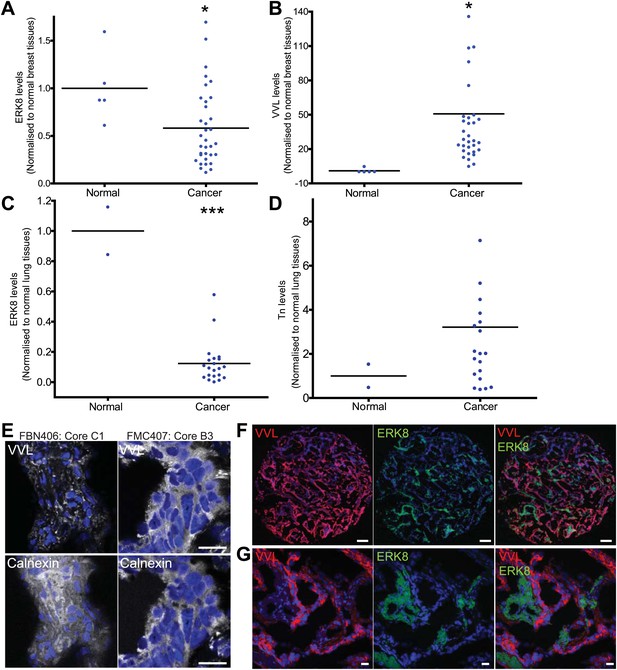
ERK8 is downregulated in human breast and lung carcinoma.
(A) Quantification of ERK8 staining in human breast biopsies. Each point represents the staining of one tissue core normalised to the average staining of the normal tissue cores. (B) Quantification of Tn (Vicia Villosa Lectin; VVL) staining in human breast biopsies. (C) Quantification of ERK8 staining in human lung biopsies. (D) Quantification of Tn staining in human lung biopsies. *p<0.05, **p<0.01, ***p<0.0001 by two-tailed unpaired t test. (E) Co-staining VVL and ER marker Calnexin revealed extensive ER co-localisation of Tn in lung carcinoma (FMC407: Core B3), whereas Tn appeared as punctuate structures in the normal lung (FBN406: Core C1). Scale bar: 20 μm. (F) ERK8 and Tn staining in a lung adenocarcinoma core (FMC407: Core B8). Scale bar: 200 μm. (G) Close-up image of the core shown in (F). Scale bar: 20 μm.
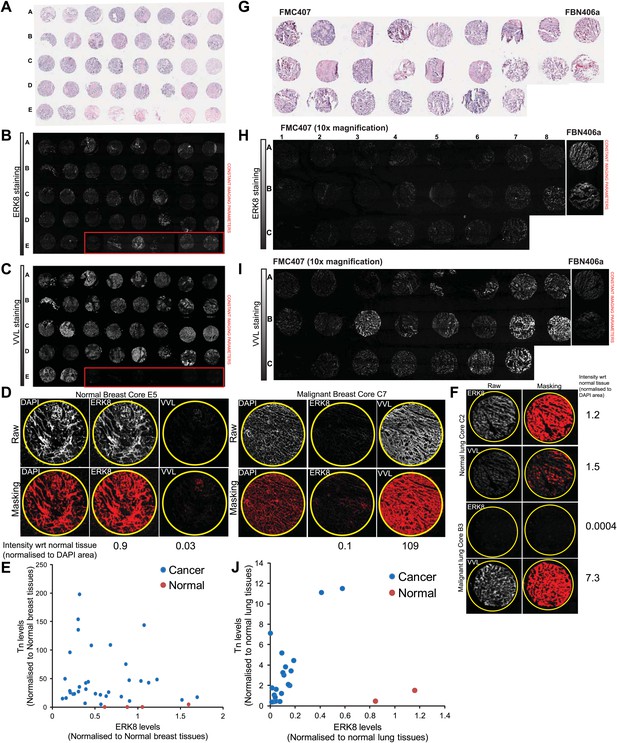
ERK8 levels are frequently reduced in human lung carcinoma.
(A) H&E staining of 40 breast biopsies on BRF404 slides purchased from US Biomax, Inc. Image is from Biomax website (http://www.biomax.us/). (B) ERK8 and (C) Vicia Villosa Lectin (VVL) were co-stained on the breast tissue cores. Red boxes indicate the positions of normal breast tissue cores. Note: Core E6 was absent on the slide, hence only 39 of the original 40 biopsies were included in the analysis. (D) Quantification method of ERK8 and VVL staining in the tissue cores. The proportion of tissue area above a fixed threshold, as highlighted by the masking, was measured. This was then normalised to the total core area represented by the nuclei (DAPI) staining. The fold change in ERK8 and VVL staining area with respect to (wrt) the average of all normal cores is presented below each example image. (E) Scatter plot of VVL and ERK8 staining in all 39 breast cores. (F) The same quantification method used in the breast array was adopted for the lung tissue array. Representative tissue core images and quantification results of the normal and cancerous lung cores are shown. (G) H&E staining of 23 lung biopsies on FBN406a and FMC407 slides purchased from US Biomax, Inc. (H) ERK8 and (I) VVL were co-stained on the lung cores. Note: Core B1 of FMC407 was absent on the slide, hence only 23 of the original 24 biopsies were included in the analysis. (J) Scatter plot of VVL and ERK8 staining in all 23 lung cores.





