Membranes linked by trans-SNARE complexes require lipids prone to non-bilayer structure for progression to fusion
Figures
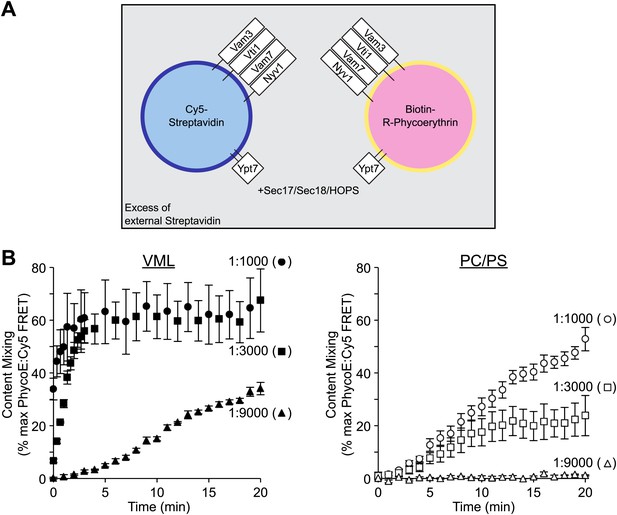
Lipid composition and SNARE concentration regulate the rate of proteoliposome membrane fusion.
(A) Membrane fusion was assayed as protected (from external non-fluorescent streptavidin) lumenal compartment mixing. This was measured as the FRET between biotin-phycoerythrin and Cy5-streptavidin, which had been entrapped within separate proteoliposome populations. Paired sets of proteoliposomes were prepared with either the complete vacuolar mixed lipids (VML) or with 70% PC/30% PS. Proteoliposomes bore Ypt7p and the 4 SNAREs, each at a 1:1000, 1:3000, or 1:9000 molar ratio to lipid phosphate, as described in the ‘Materials and methods’. For each pair, half the proteoliposomes had 0.3% of its lipid as Marina Blue-PE and bore lumenal Cy5-streptavidin, while the complementary proteoliposomes had 1.5% NBD-PE and bore lumenal biotinylated-phycoerythrin. (B) Fusion assays were performed with Sec17p, Sec18p, HOPS, and ATP in the presence of excess nonfluorescent streptavidin, as described in the ‘Materials and methods’. Error bars here and in subsequent figures are the standard deviations from three assays.
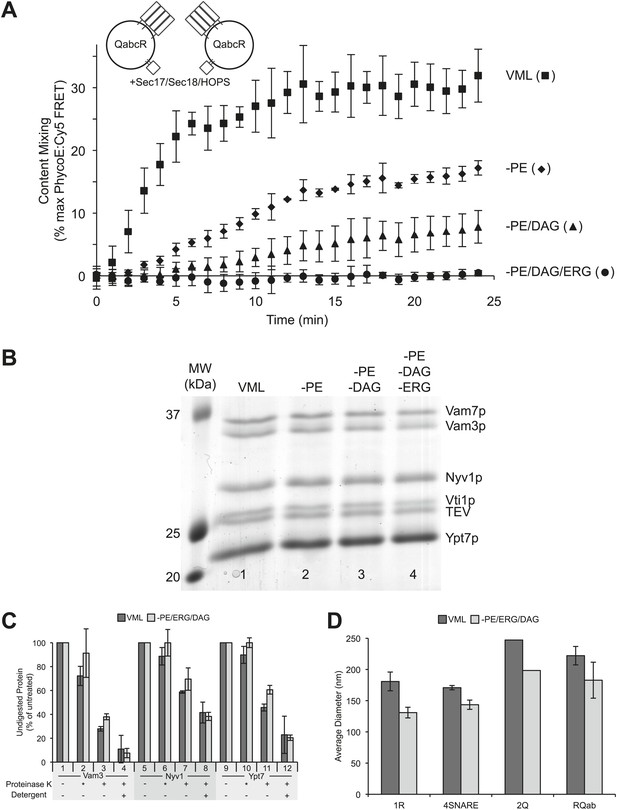
A role for neutral, non-bilayer lipids in fusion.
(A) Fusion of proteoliposomes bearing Ypt7p and 4-SNAREs (1:5000 molar ratio to lipid phosphate), prepared with either the complete vacuole lipid mix (squares) or missing PE (diamonds), PE and DAG (triangles), or PE, DAG, and ERG (circles). Fusion was assayed as the FRET between lumenal fluorescent proteins in the continuous presence of an excess of external nonfluorescent streptavidin. (B) Proteoliposomes were analyzed for their protein composition by SDS-PAGE and Coomassie blue staining. Lipid composition: Lane 1, complete vacuolar lipid mix; lane 2, PE omitted; lane 3, PE and DAG omitted; lane 4, PE, DAG, and ERG omitted. In each case, the percentage of PC was increased to account for the omitted lipid(s). (C) Similar protease-accessibility of SNAREs and Rab across proteoliposomes preparations. Proteoliposomes (1.2 mM lipid) were incubated in 15 µl RB150 with 60 mM HEPES/NaOH pH 8.0 for 10 min at 27°C with either no addition of protease, with 60 µg/ml of proteinase K which had been preincubated for 10 min with 1 mM PMSF prior to proteoliposome addition (indicated by asterisk), with fully-active proteinase K which had not been preincubated with PMSF, or with fully active proteinase K and 1% (wt/vol) β-octylglucoside. After this incubation, PMSF was added to samples which had fully active proteinase K and the incubation continued for an additional 10 min. All samples were then mixed with SDS sample buffer, heated to 95°C for 5 min and subjected to SDS-PAGE. Gels were stained with Coomassie blue, and bands corresponding to Vam3p, Nyv1p, and Ypt7p quantified by scanning with a Microtek Bio-5000 scanner (Microtek Lab, Inc., Santa Fe Springs, CA) and UN-SCAN-IT gel 5.3 software (Silk Scientific, Orem, UT). For each of these three proteins, the intensity of the band from samples which never saw proteinase K was set to 100%. Dark bars correspond to VML proteoliposomes, light bars to proteoliposomes prepared without PE, DAG, and Erg. Shown is the average of two experiments +/− standard deviations. (D) The size distribution of various proteoliposome preparations was analyzed by dynamic light scattering with a Zetasizer nano ZS (Malvern Instruments Inc., Westborough, MA) through non-invasive back-scatter at 173°. For each liposome preparation, at least four samples (400 µl at a lipid concentration of 20 µM) were measured in low volume disposable sizing cuvettes at 25°C. Shown is the average diameter (+/− standard deviation) of independent proteoliposome preparations composed of the complete vacuolar lipid mix (dark bars) or without PE, ERG, DAG (light bars) and bearing either Nyv1 (1R; n = 3), all four SNAREs (4SNARE; n = 2), Vam3 and Vti1 (2Q; n = 1), or Nyv1, Vam3, and Vti1 (RQab; n = 2).
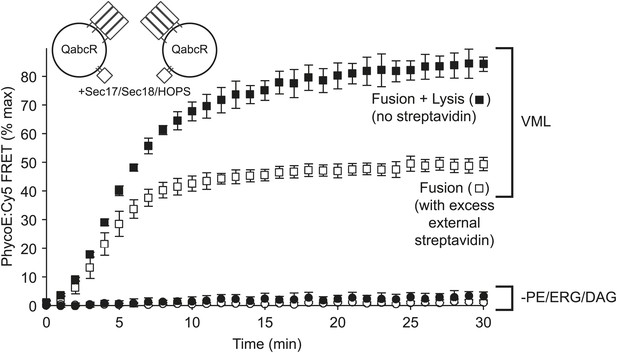
Omission of PE, DAG, and ERG blocks HOPS, Sec17p, and Sec18p triggered lysis as well as fusion.
Proteoliposomes of complete vacuolar lipid mix or lacking PE, DAG, and ERG and with Ypt7p and the 4 vacuolar SNAREs (1:5000 molar ratio to lipid phosphate) were incubated either with a large molar excess of non-fluorescent streptavidin, restricting FRET development to sealed fusion events, or without external streptavidin, yielding FRET from both fusion and from lysis (Zucchi and Zick, 2011).
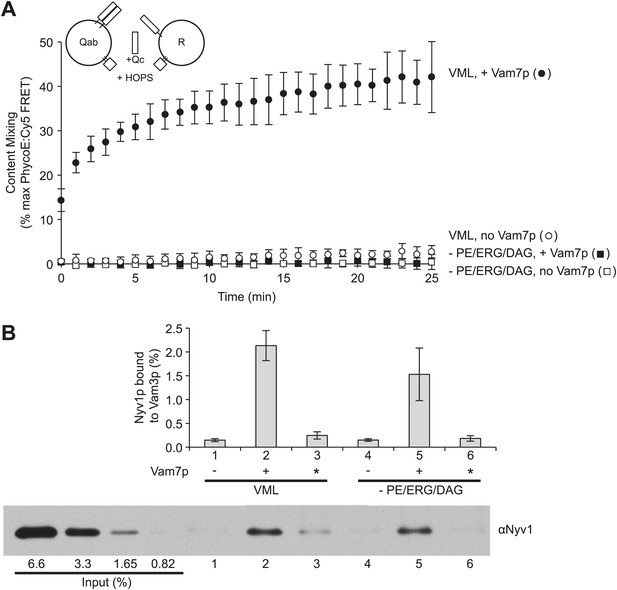
Small-headgroup, nonbilayer lipids are needed for trans-SNARE docked membranes to proceed to fusion.
Reconstituted proteoliposomes with either the R-SNARE or the Vam3p and Vti1p Q-SNAREs, prepared at a 1:5000 molar ratio of SNARE to lipid and either having the complete vacuolar lipid mix or without PE, ERG, or DAG were incubated in fusion reactions. Vam7p (0.5 µM) was added where indicated, either during the fusion reaction (lanes 2 and 5) or after the reaction was terminated by detergent addition (indicated by an asterisk, lanes 3 and 6). Each reaction was (A) assayed for lumenal content mixing and (B) mixed after 10 min with a 10-fold volume of a modified RIPA buffer (20 mM HEPES/NaOH, pH 7.4, 0.15M NaCl, 0.2% bovine serum albumin (defatted), 1% Triton X-100, 1% sodium cholate, 0.1% sodium dodecyl sulfate, 1 mM EDTA) with 40 µg/ml affinity-purified antibody to Vam3p and 1 µM recombinant soluble domain of Snc2p to suppress SNARE complex assembly in detergent. After addition of 10 µl of RIPA buffer-washed suspension of magnetic beads with bound protein A (Thermo Scientific), samples were mixed for 1 hr at room temperature. Beads were collected by placing the tubes for 2 min onto a magnetic rack, and the unbound proteins removed. Beads were thrice washed with 1 ml of modified RIPA buffer, then proteins were eluted with SDS sample buffer at 95°C and analyzed by SDS-PAGE and immunoblot with antibodies to Nyv1p. Reactions were performed without further SNARE addition, with 0.5 µM Vam7p from the start of the incubation, or with the Vam7p added one minute after solubilization by RIPA buffer. The same preparations and solutions were premixed, then used in parallel for the assays of fusion and trans-assembly of SNAREs shown here. The immunoblot of one of the three trans-SNARE assays is shown.
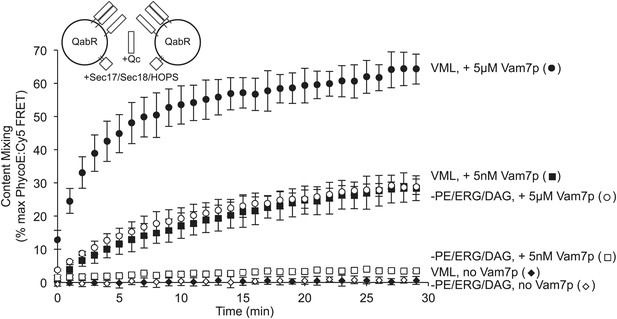
The requirement for PE, ERG, and DAG is governed by the level of trans-SNARE complex.
Fusion assay pairs of proteoliposomes were prepared with Ypt7p, with VML lipids or lacking PE, ERG, and DAG, as indicated, and with Nyv1p, Vam3p, and Vti1p (the R- and Qa- and Qb-SNAREs, respectively) at a 1:1000 molar ratio to total lipid. Fusion assays were initiated by the addition of HOPS, Sec17p, and Sec18p, as described in the ‘Materials and methods’, as well as the indicated (final) concentration of Vam7p.
Tables
Protein abundance, relative to lipids, in vacuoles or reconstituted proteoliposomes (RPL) fusion reactions
| Protein | Molar ratio of lipid:protein in RPL reactions* | Molar ratio of lipid:protein on vacuoles | Ratio (RPLs/vacuoles) of molar protein:lipid ratios in std. reactions† | |
|---|---|---|---|---|
| BJ3505 | DKY6218 | |||
| Vam7p | 2 × 103 | 30 × 104 | 6.5 × 104 | 7 × 101 |
| Vam3p | 2 × 103 | 11 × 104 | 22 × 104 | 7 × 101 |
| Vti1p | 2 × 103 | 10 × 104 | 13 × 104 | 5 × 101 |
| Nyv1p | 2 × 103 | 4.3 × 104 | 8.1 × 104 | 3 × 101 |
| Ypt7p | 4 × 103 | 1.9 × 104 | 1.8 × 104 | 0.5 × 101 |
| Sec17p | 7 × 103 | 41 × 104 | 13 × 104 | 3 × 101 |
| Sec18p | 1 × 103 | 10 × 104 | 13 × 104 | 10 × 101 |
| Vps33p | 6 × 103 | 17 × 104 | 31 × 104 | 3 × 101 |
-
*
for SNAREs and Ypt7p, calculated, based on a 1:1000 lipid:protein ratio during reconstitution and an assumption of 50% outwardly-oriented SNAREs on proteoliposomes; for others, based on amounts of added proteins, and 0.74 mM lipids in standard proteoliposome reactions (see ‘Materials and methods’).
-
†
based on measured (see ‘Materials and methods’) values of 2.17 nmol lipid per µg total vacuole protein for BJ3505 vacuoles and 1.00 nmol lipid per μg total vacuole protein for DKY6218 vacuoles, and standard vacuole reactions containing 3 µg protein of each vacuole in 30 µl.





