Evolution of insect olfactory receptors
Figures
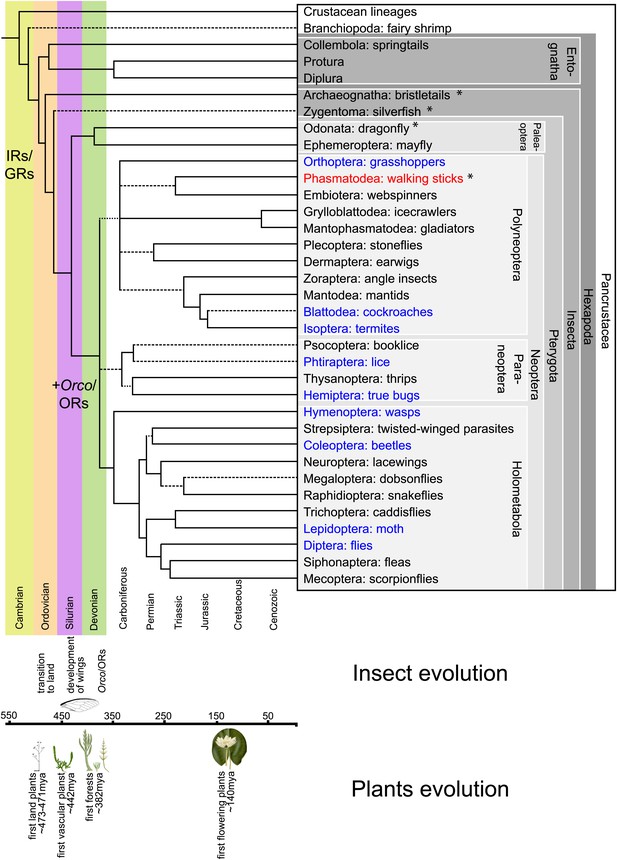
Hexapodan phylogeny.
Phylogeny was adapted from Trautwein et al. (2012). Timescale was adjusted for higher level taxa based on Rota-Stabelli et al. (2013), for Holometabola according to Wiegmann et al. (2009) and the remaining groups based on their fossil record (http://insects.about.com/od/evolution/a/Timeline-of-Fossil-Insects-by-Order.htm), in order to correlate important events in plant and insect evolution with the emergence of insect olfactory receptors. IRs and GRs are known to be much older than insects (Peñalva-Arana et al., 2009; Croset et al., 2010), however, ORs and Orco have evolved during the evolution of insects and cannot be found outside the insect clade (Peñalva-Arana et al., 2009). Insects with a described OR/Orco-based olfactory system were highlighted in blue, whereas species were Orco was described in this study were colored in red. All orders investigated in this study are labeled by an asterisk. Our data suggests the evolution of the coreceptor Orco after the bristletails split from its last common ancestor with the remaining insects. However, an olfactory system that relies both on ORs and Orco seems to have evolved after the emergence of wings.
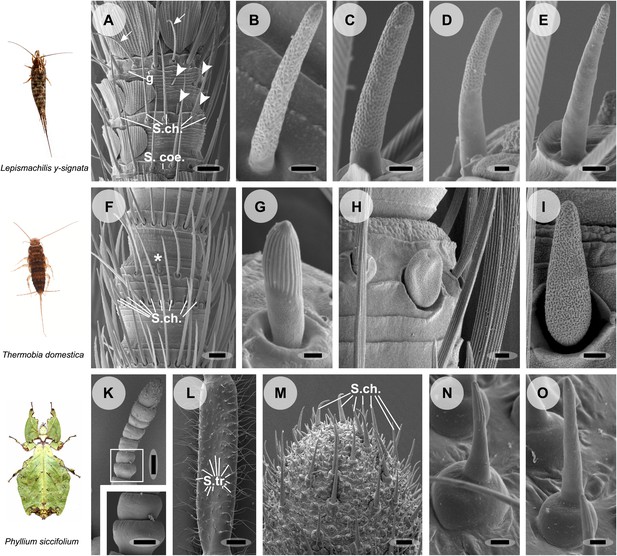
Olfactory sensilla on the antennae of L. y-signata (A–E), T. domestica (F–I) and P. siccifolium (K–O).
Animals are depicted next to the corresponding antennal SEM images. (A) Detailed view of the antennae of L. y-signata. The proximal part of the antennae is not only covered with sensilla, but also scales. Glands (g) are highly abundant on the antennae. Many mechanosensory sensilla (S.ch.: Sensilla chaetica) were arranged in circles on the antennal segments. On some antennal segments gustatory sensilla (arrows) can be found between the S.ch (for further information read Missbach et al., 2011). Very rarely zero to four olfactory Sensilla basiconica were identified per segment, in a mostly redundant pattern on the antennae with similar numbers of olfactory sensilla and sensilla types on each antennal segment. Antennal segments are separated by antennal breaking points. The pattern of sensilla is modulated by increasing the number of annuli of a segment through molting. (B–E) Different morphological types of basiconic sensilla. No grooved sensilla/olfactory coeloconic sensilla were identified on the antennae. Only small pegs surrounded by a cuticular wall (s. coe.; referred as coeloconica-like sensillum, Bockhorst 1988) were located on the antennae. These sensilla are not olfactory (for detailed external morphology see Missbach et al., 2011). (F) Detailed view of the antennae of T. domestica. The antennal organization is similar to the bristletail, with antennal breaking points and lifelong molting. The most abundant sensilla on the antennae again are mechanosensory S.ch.; beside those gustatory and olfactory sensilla are distributed in a species-specific modular manner over the antennae. (G) In contrast to L. y-signata, grooved sensilla can be found on the antennae of T. domestica. (H and I) Different morphological types of basiconic sensilla. (K and L) Gender specific differences between a female (K) and a male (L) antennae of P. siccifolium. Female antennae are short and lack trichoid sensilla (S.tri.). They more or less lack sensilla on the proximal annuli, only the last two annuli are covered with a high number of olfactory and also some mechanosensory sensilla (S.ch.). (M) Male antennal tip. Similar to the distal female antennal annuli the highest density of sensilla can be found on the last annuli. (N and O) Both grooved and pored sensilla can be found on these segments. Scale bars: A: 50 μm; B, C, D, E, H, I, N, O: 2 μm; F: 100 μm; G: 1 μm; K, L: 200 μm; M: 20 μm.
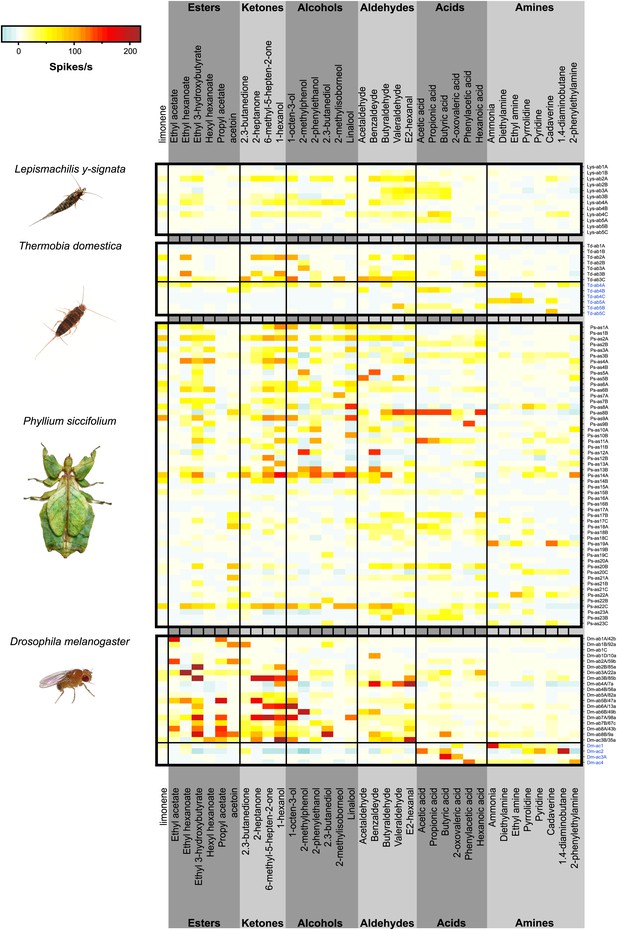
Color coded response profiles of L. y-signata, T. domestica, P. siccifolium and D. melanogaster.
Spikes are sorted by neurons, with the exception of ac1, ac2, and ac4 of D. melanogaster where spike sorting was not possible. Means over 5 to 23 recordings were used as basis for visualization (source data are given in Figure 3—source data 1). The same color code was used for all species, ranging from highest to lowest encountered change in activity. Neurons in grooved sensilla are indicated by blue letters (ac). For L. y-signata responses to odors were only obtained from neurons in porous sensilla (ab). A separation between porous and grooved sensilla was not possible for P. siccifolium. Sensilla were classified as antennal sensillum (as). L. y-signata neurons are mostly broadly tuned with comparable low change in spiking activity. For P. siccifolium a total of 23 different functional sensillum types were identified in SSR recordings (in comparison five in L. y-signata, five in T. domestica) suggesting that leaf insects have a broader response repertoire.
-
Figure 3—source data 1
Excel file of mean responses and baseline firing rate of the different OSN classes of L.y-signata, T. domestica, P.siccifolium, and D. melanogaster.
- https://doi.org/10.7554/eLife.02115.006
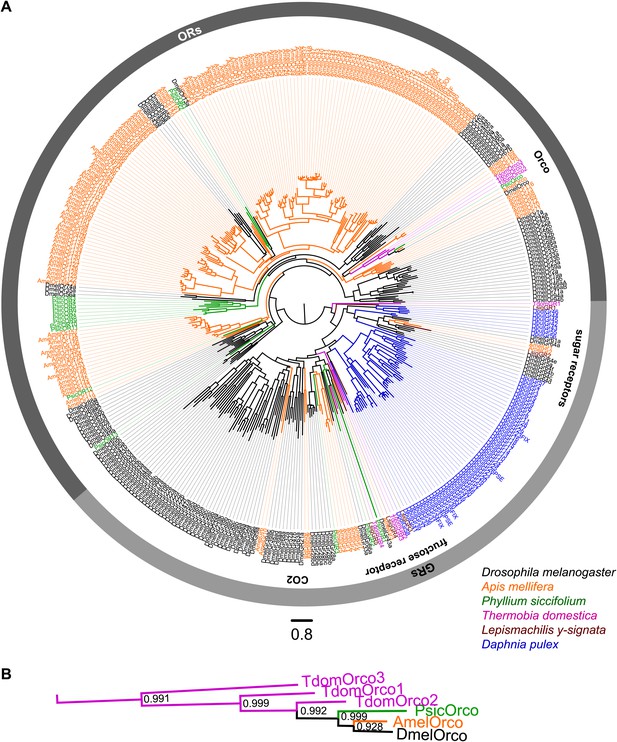
ORs and GRs of L. y-signata, T. domestica, and P. siccifolium.
(A) Dendrogram displaying the relationship of identified OR and GR candidates of L. y-signata, T. domestica, and P. siccifolium to D. melanogaster (Clyne et al., 1999; Gao and Chess, 1999; Vosshall et al., 1999) and Apis mellifera (Robertson and Wanner, 2006) GR and OR proteins, and Daphnia pulex GRs (Peñalva-Arana et al., 2009). The dendrogram was determined by maximum likelihood analysis of a MAFFT-Alignment using FastTree2. All L. y- signata candidates group within the GRs. Only candidates with a translated amino acid sequence longer than 120 amino acids and overlap in multiple sequence alignment were taken for analysis, since ORs and GRs are highly divergent and only unigenes should be included in the analysis (all candidate OR and GR sequences of L. y-signata, T. domestica and P. siccifolium are given in Figure 4—source data 1 for amino acids and Figure 4—source data 2 for nucleotide sequences). For T. domestica, we identified three different variant Orco types that were included in the analysis as full length translated amino acid sequences. (B) Blow-up of the dendrogram showing the support values for the coreceptor subgroup. The whole group is well supported.
-
Figure 4—source data 1
Amino acid sequences of putative olfactory and gustatory receptors of L. y-signata, T. domestica, and P. siccifolium.
- https://doi.org/10.7554/eLife.02115.010
-
Figure 4—source data 2
Nucleotide sequences of putative olfactory and gustatory receptors of L. y-signata, T. domestica, and P. siccifolium.
- https://doi.org/10.7554/eLife.02115.011
-
Figure 4—source data 3
MAFFT-alignment of OR and GR candidates of L. y-signata, T. domestica, P. siccifolium and D. melanogaster (Clyne et al., 1999, Gao and Chess, 1999, Vosshall et al., 1999) and Apis mellifera (Robertson and Wanner, 2006) GR and OR proteins, as well as Daphnia pulex GRs done.
- https://doi.org/10.7554/eLife.02115.012
-
Figure 4—source data 4
FastTree file resulting from the MSA of Figure 4—source data 3 (can be opened with FigTree).
- https://doi.org/10.7554/eLife.02115.013
-
Figure 4—source data 5
Tree file resulting from the MSA of Figure 4—source data 3 containing node support values (can be opened e.g., with Adobe Illustrator).
- https://doi.org/10.7554/eLife.02115.014
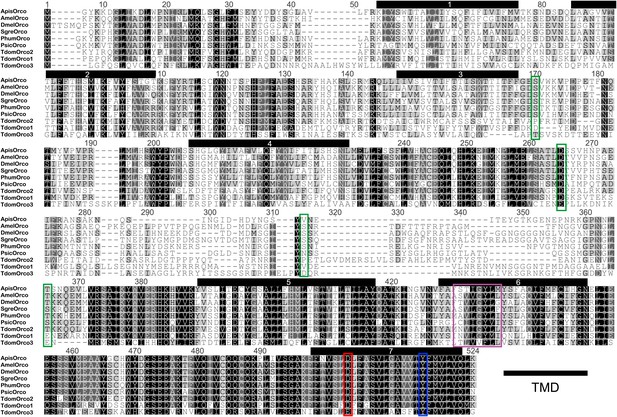
Multiple sequence alignment of T. domestica Orcos.
Alignment of T. domestica Orcos with Orcos of Acyrthosiphon pisum (GI:328723530), A. mellifera (GI:201023349), D. melanogaster (GI:24644231), Schistocerca gregaria (GI:371444780), Pediculus humanus corporis (GI:242009783), P. siccifolium (this study). Important amino acids are highlighted in colored boxes (purple: effect on ion permeability, Wicher et al., 2008; green: phosophorylation sites for PKC of DmelOrco, Sargsyan et al., 2010; blue: affect spontaneous and evoked action potentials in receptor complex, Nakagawa et al., 2012; red: important residue for channel activity, Kumar et al., 2013).
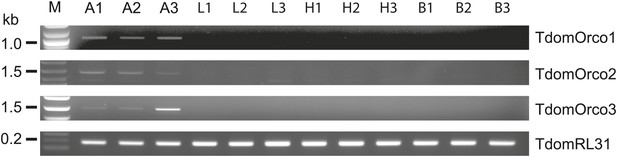
Expression of T. domestica Orcos.
Using RT-PCR Orco expression was detected in the antennae (A) of T. domestica, but not in legs (L), heads without antennae and palps (H), and bodies (B). Primer sequences are given in Figure 6—source data 1.
-
Figure 6—source data 1
Primers and their properties used in this study.
- https://doi.org/10.7554/eLife.02115.017
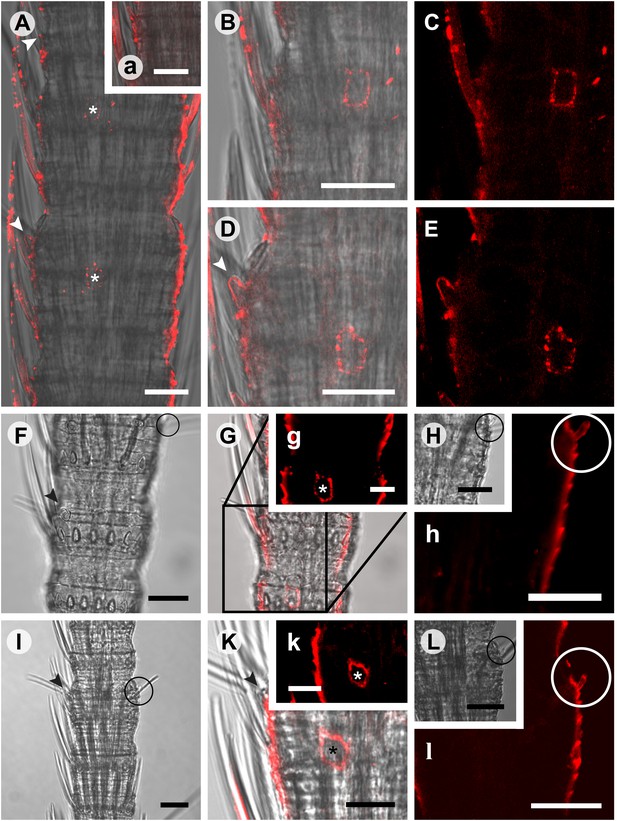
In situ hybridization on whole mount antennae of T. domestica using a Dig-labeled TdomOrco1 antisense probe.
(A) Part of a T. domestica antenna. Combined image of fluorescent and transmitted light channel taken with cLSM. The positions of pored sensilla are indicated by arrowheads, with the upper sensillum displayed in the small box in the upper right corner. Labeled cell bodies are assigned by asterisks. (B–E) Single confocal planes through the antenna. Only a single soma close to each pored sensillum is labeled suggesting that only one neuron per sensillum expresses this Orco variant. In B and D some precipitate is visible. (C and E) Same image section as B and D, but without transmitted light. (F) Transmitted light image of a part of a second antenna. Location of a pored sensillum is again assigned by an arrowhead. A grooved sensillum indicated by a black circle is situated on the opposite side of the antenna. (G) Same part of the antenna taken with transmitted light and fluorescent channel. Again only one soma is labeled close to a pored sensillum. g: Only the Dig signal. Cuticle shows a strong autofluorescence on both sides. H, h: No signal was obtained close to a grooved sensillum. (I) Part of another antenna with a pored and a grooved sensillum on the same annulus. K, k: Image section from the part of the antenna close to the pored sensillum. A single soma is labeled by the probe. k: Only the fluorescent signal. L, l: No soma was labeled close to the grooved sensillum. For sense controls view Figure 7—figure supplement 1. Scale bars A–F, H, I, L: 20 μm; g, K, k: 10 μm.
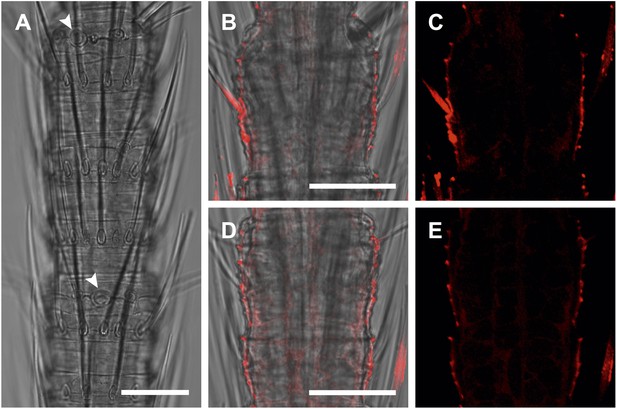
In situ hybridization on the antenna of T. domestica using sense probes directed against the TdomOrco1.
(A) Transmitted light images taken with cLSM. The position of olfactory sensilla is indicated by arrowheads. (B–E) Projection section through the antennae. No Dig signals were obtained using the sense probes of TdomOrco1. All scale bars 20 μm.
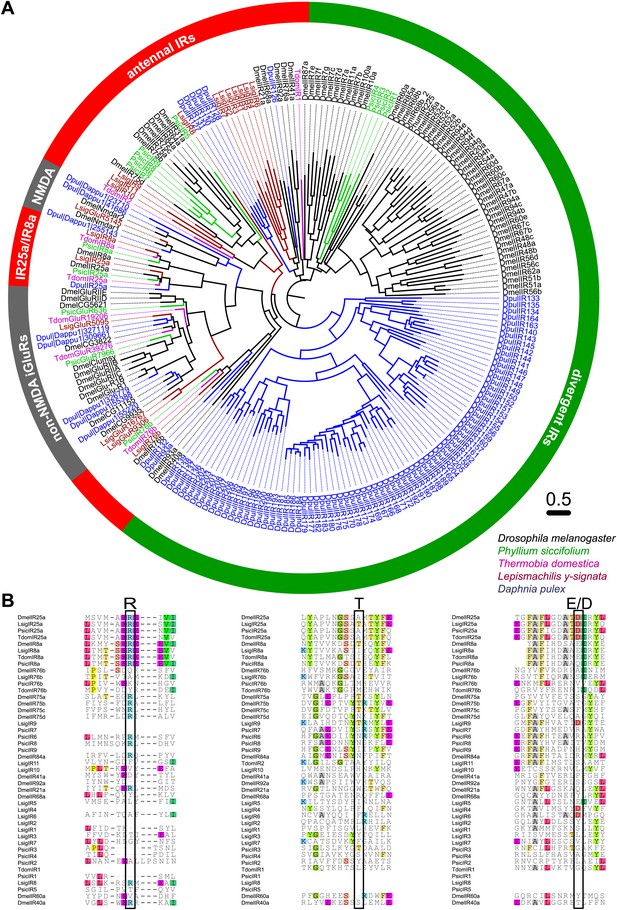
Ionotropic glutamate receptors of L. y-signata, T. domestica, and P. siccifolium.
(A) Analysis of the relationship between L. y-signata, T. domestica, P. siccifolium, D. melanogaster and D. pulex iGluRs and IRs (D. melanogaster and D. pulex sequences were sequences taken from Croset et al., 2010). Amino acid sequences were aligned using the MAFFT alignment tool plug-in in Geneious Pro 5.0.4 (BLOSUM72, gap open penalty: 1.53, offset value: 0.123, E-INS-i settings). The dendrogram was generated using maximum likelihood analysis with FastTree2. (All candidate IR sequences of L. y-signata, T. domestica, and P. siccifolium are given in Figure 8—source data 1 for amino acids and Figure 8—source data 2 for nucleotide sequences) (B) Excerpts of the alignment showing the predicted glutamate binding domains and key amino acids. Mutations in one or several of the key amino acids are a structural feature to distinguish between iGluRs and IRs, although they can be present in the coreceptors.
-
Figure 8—source data 1
Amino acid sequences of putative variant ionotropic glutamate receptors of L. y-signata, T. domestica, and P. siccifolium.
- https://doi.org/10.7554/eLife.02115.021
-
Figure 8—source data 2
Nucleotide sequences of putative variant ionotropic glutamate receptors of L. y-signata, T. domestica, and P. siccifolium.
- https://doi.org/10.7554/eLife.02115.022
-
Figure 8—source data 3
MAFFT amino acid alignment of iGluR and IR candidates of L. y-signata, T. domestica, P. siccifolium, D. melanogaster, and D. pulex (D. melanogaster and D. pulex sequences were sequences taken from Croset et al., 2010).
- https://doi.org/10.7554/eLife.02115.023
-
Figure 8—source data 4
FastTree file resulting from the MSA of Figure 4—source data 3 (can be opened with FigTree).
- https://doi.org/10.7554/eLife.02115.024
-
Figure 8—source data 5
Tree file resulting from the MSA of Figure 8—source data 3 containing node support values.
- https://doi.org/10.7554/eLife.02115.025
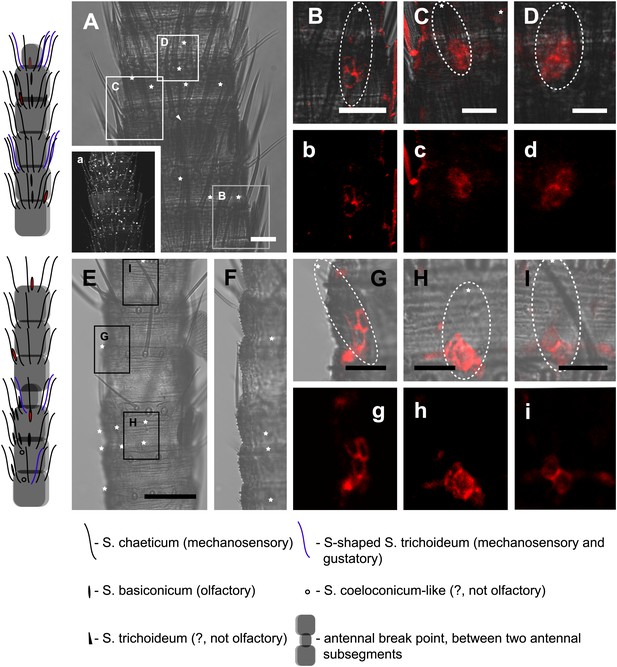
In situ hybridization on the antenna of L. y-signata using a Dig-labeled LsigIR25a antisense probe.
On the left: schematic drawings of the position of the different sensillum types on the particular antennal subsegment. The legend for the sensillum types is given below the confocal images. (A–D) Labeling of somata in a subsegment of an antenna. Mostly two somata were labeled with the probe. The positions of the somata were in line with the positions of basiconic sensilla, but not gustatory and mechanosensory sensilla. Ultrastructural investigation of basiconic sensilla of Machilis sp. (Archaeognatha) and Lepisma saccharina (Zygentoma) suggests that the sensory neurons are located in a distance of at least 25 μm from the sensillum base in the extension of the sensillum (Berg and Schmidt, 1997). Therefore, we concluded that the labeled somata correspond to neurons housed in basiconic sensilla. These sensilla were colored red in the drawing on the left. (A) Transmitted light overview with asterisks labeling basiconic sensilla. Image sections given in B–D are indicated by white boxes and the corresponding letters. a: Projection of confocal planes recorded with Argon laser at a wavelength of 488 nm to identify the position of basiconic sensilla. (B–D) Overlaid transmitted light and fluorescent images of labeled somata. b–d: Images without transmitted light channel. (E–I) Labeling of somata in a second antenna. Parts of two antennal segments that are separated by an antennal break point. The break point can be recognized by a thinner segment on the distal part of the antennae or by a special trichoid sensillum that is only present on the segment proximal to a breaking point. (E and F) Transmitted light images of the antenna. E is more from the top. Image sections given in G–I are indicated by white boxes and the corresponding letters. F is more central plane. Asterisks denote the location of a basiconic sensillum. (G–I) Overlaid confocal images of labeled neurons. Images are projections of three confocal planes. On some positions the cuticle is given a background signal. g–i: Images without transmitted light channel. Scale bars: A–C, G–I: 20 μm, E: 50 μm, D: 10 μm.
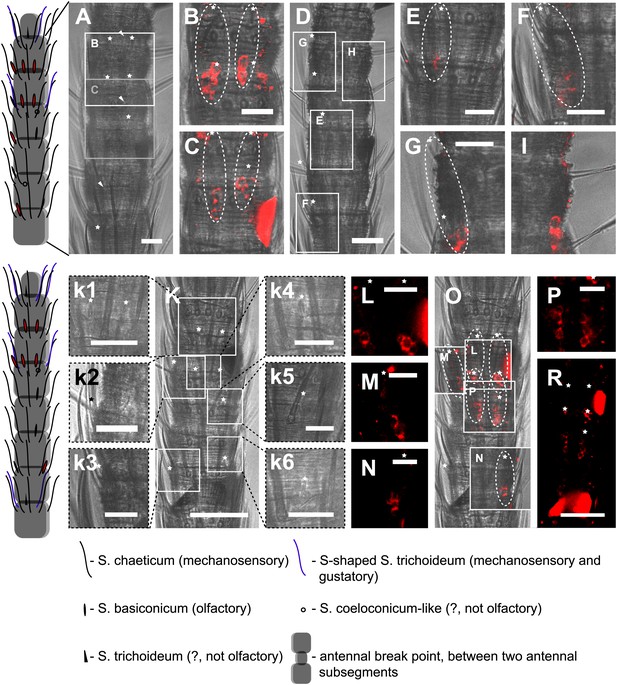
In situ hybridization on the antenna of L. y-signata using an antisense probe directed against the IR coreceptor IR8a.
On the left: schematic drawings of the position of the different sensillum types on antennae. The legend for the sensillum types is given below the confocal images. (A–I) Labeling of somata in a subsegment of one antenna, with A and D showing the position of basiconic sensilla in the transmitted light channel. Images that are shown in B, C and E–I are indicated by white boxes and the corresponding letters. (B, C, E–I) Overlaid transmitted light and fluorescent images of single confocal planes through the dorsal side of the antenna. The signals are lineal to basiconic sensilla in a distance of about 20–25 μm to the base of the sensillum. Therefore, we conclude that the labeled somata correspond to basiconic sensilla. For the somata in I, we could not find a corresponding sensillum. It might be situated on the backside of the antenna. Red fluorescence on the sides is due to autofluorescence of the antennal cuticle. (K–R) Labeling of somata in a second antenna. (K, k1–k4) Transmitted light images of different focal planes, giving an overview about the position of basiconic sensilla on this antennal subsegment. (O) Merged image of transmitted light image and overlaid confocal and transmitted light image. Detailed images of the signals within the white boxes are displayed with corresponding letters around the overview image (L–N, P). (R) Single optical section through the antenna. Background fluorescence according to antennal cuticle and sensillum bases made whole antennal projections not possible. Very pronounced on this picture are the antennal scales on the right side of the antenna and the bottom of the picture. Scale bars: A–I, k1–k6, L–N, P: 20 μm, K, R: 50 μm.
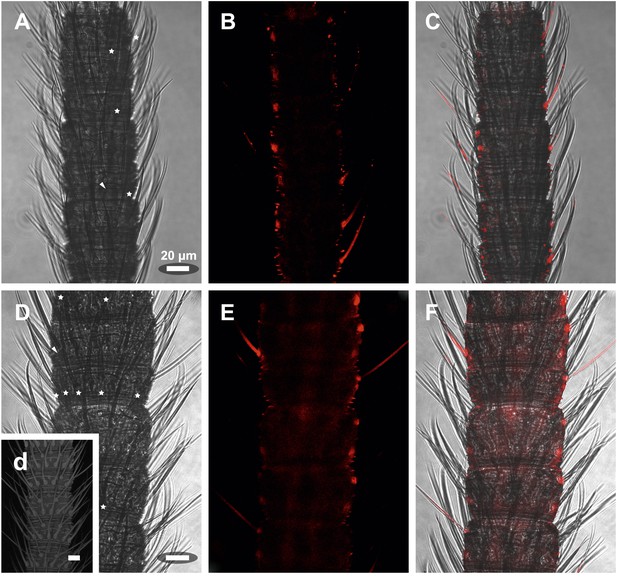
In situ hybridization on the antenna of L. y-signata using sense probes directed against the IR coreceptors IR25a, IR8a.
(A, D, G) Transmitted light pictures of antennal excerpts. Asterisks mark positions of basiconic sensilla, arrowheads places were coeloconic-like sensilla are located. No signals were obtained in the sense controls. (A–C) IR25a sense probe. (D–F) IR8a sense probe. All scale bars 20 µm.
Tables
Technical overview of transcriptomes (study accession: PRJEB5093, study unique name: ena-STUDY-MPICE-12-12-2013-15:03:23:860-31)
| Organism | Sequencing technique | Number of reads | Number of contigs above 400 bp | N50 | Average length of contigs | Tissue | Sample accession | Secundary accession | Sample unique name |
|---|---|---|---|---|---|---|---|---|---|
| Lepismachilis y-signata | HiSeq2000 (Illumina) | 22’444’128 | 68’984 | 1’179 | 1’000 | antennae and palps | ERS384175 | SAMEA2276780 | Lysig1 |
| HiSeq2500 (Illumina) | 77’060’687 paired end | antennae | ERS384176 | SAMEA2276781 | Lysig2 | ||||
| HiSeq2000 (Illumina) | 25’242’666 | 37’860 | 857 | heads, whole bodies | ERS399748 | SAMEA2342071 | LysigMix1 | ||
| Thermobia domestica | HiSeq2500 (Illumina) | 27’704’231 paired end | 31’172 | 1’349 | 1’070 | antennae | ERS384177 | SAMEA2276782 | Tdom1 |
| Phyllium siccifolium | HiSeq2500 (Illumina) | 30’762’777 paired end | 34’653 | 1’890 | 1’305 | antennae | ERS384178 | SAMEA2276783 | Psic1 |
Number of candidate contigs (not unigenes) for the different gene families identified in the transcriptomes of the different species
| Organism | Orco | ORs | GRs | IRs |
|---|---|---|---|---|
| Lepismachilis y-signata | – | – | 7 (5 above 400 bp) | 17 (16 above 400 bp) |
| Thermobia domestica | 6 (1 above 400 bp) | – | 9 (3 above 400 bp) | 19 (9 above 400 bp) |
| Phyllium siccifolium | 1 (1 above 400 bp) | 30 (16 above 400 bp) | 6 (2 above 400 bp) | 32 (19 above 400 bp) |





