Science Forum: Considerations when investigating lncRNA function in vivo
Figures
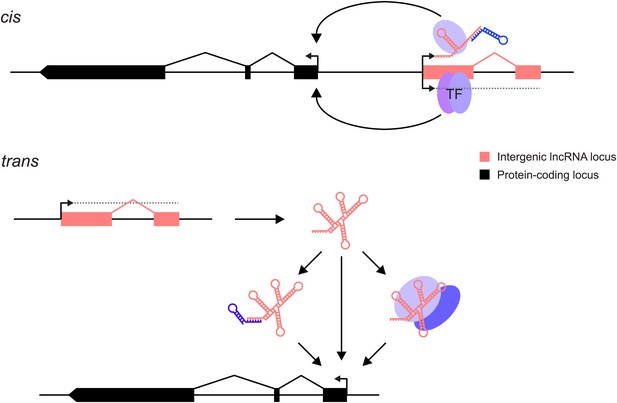
lncRNAs can act through cis and/or trans mechanisms. lncRNAs (pink) can act to regulate expression of their genomically neighbouring protein-coding genes (black) in cis (upper panel), or of distant protein-coding genes in trans (lower panel).
In both situations, the RNA moiety itself may act through binding to cellular proteins (blue ovals) or via base-pairing with other RNAs (blue stem-loop) to modulate their function or binding. The RNA may also directly bind double-stranded DNA in trans (Grote et al., 2013) or in cis (Senner et al., 2011). The lncRNA locus (pink) may also encompass transcription factor binding sites (TF) that regulate the transcription of neighbouring genes. This effect may either be entirely independent of the lncRNA, or the binding of transcription factors may be affected positively or negatively by the act of transcription through the lncRNA locus. In this case, the mature RNA product would be incidental.
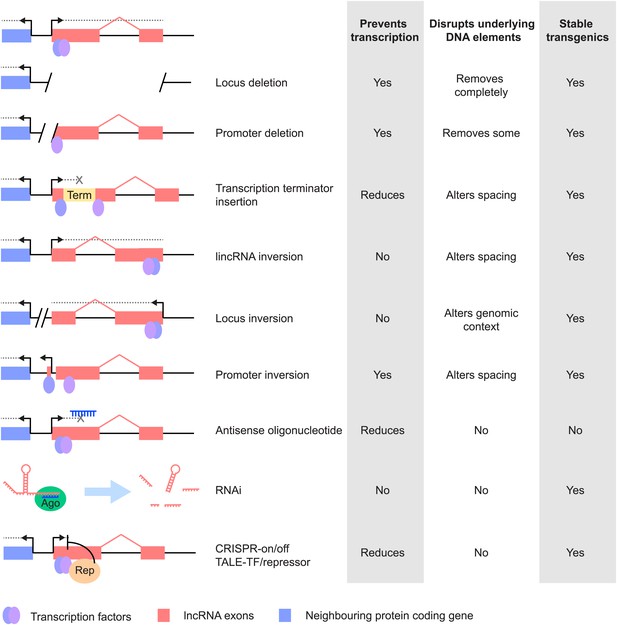
Different strategies for analysis of lncRNA loss-of-function. Strategies that have been used to alter lncRNA function are described pictorially, with the wild type situation on the top-most line.
The lncRNA locus is indicated in pink, neighbouring protein-coding gene in blue, transcription factor binding sites within it by blue and purple ovals, transcriptional terminator sequences in yellow (‘Term’) and the process of transcription by grey dotted lines. Antisense oligonucleotides are able to bind to nascent RNA transcripts and trigger RNase H mediated degradation of the transcript in the nucleus. RNAi is elicited by short RNA species that bind to argonaute proteins (Ago, green oval) within the cell. This complex recognises complementary lncRNA molecules in the cytoplasm, and triggers their destabilisation by the endogenous cellular machinery. The CRISPR and TALE systems use designer DNA binding factors to recruit repressor or activator domains (orange oval) to the lncRNA to affect transcriptional initiation. The effects of each strategy upon the process of transcription and presence of underlying DNA elements such as transcription factor binding sites are indicated. The possibility of generating stable transgenic animals to investigate phenotypes throughout development is also noted.
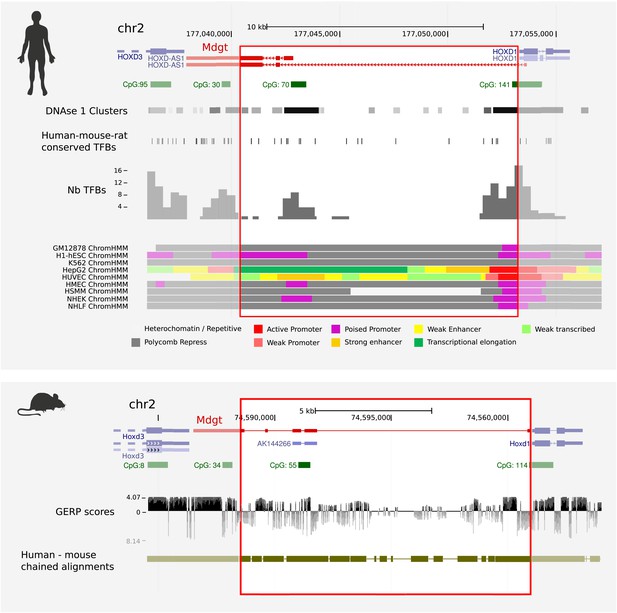
Human and mouse ENCODE data indicate that Mdgt−/− lines contain deletions of conserved binding sites for transcription factors and chromatin regulatory proteins.
The engineered deletion in mouse, and its equivalent sequence in human, are indicated by red rectangles, and spans 85% (12.4 kb of 14.7 kb) of intergenic sequence between mouse Hoxd1 and Hoxd3. Mdgt, virtually shares its start site with Hoxd1, a gene expressed with exquisite specificities in only a few cell populations during early development (Zakany et al., 2001). Predicted transcription factor binding sites (TFBs) that are conserved in human, mouse and rat are shown against the human genome (Consortium, 2012; Ernst and Kellis, 2012). Numbers of experimentally-determined TFBs per genomic interval are shown in the histogram, and clusters of DNase 1 hypersensitivity sites, are also shown aligned against the human locus. Predicted CpG islands acquired from the UCSC Genome Browser are shown in green, and chained human-mouse alignments are shown in olive green. Evolutionary conservation (GERP) scores are indicated below the mouse locus.
Tables
Representative studies that have disrupted lncRNA loci in vivo (N/A—not applicable)
| lncRNA name | Organism | Mutation strategy | Reported animal phenotype | RNA-based rescue? | Reference |
|---|---|---|---|---|---|
| Xist | Mus musculus | ∼15 kb replaced with a neo expression cassette | Females inheriting paternal allele were embryonic lethal; males fully viable | No | (Marahrens et al., 1997) |
| Xist | Mus musculus | Inversion of Exon 1 to intron 5 | Embryonic lethality of paternally inherited allele | No | (Senner et al., 2011) |
| H19 | Mus musculus | Replacement by neo cassette | Slightly increased growth | No | (Ripoche et al., 1997) |
| roX | Drosophila melanogaster | Deletions of roX1 or roX2 | None, except when in combination: male‐specific reduction in viability | Yes | (Meller and Rattner, 2002) |
| Kcnq1ot1 | Mus musculus | Promoter deletion | Growth deficiency for paternally inherited mutation | No | (Fitzpatrick et al., 2002) |
| Airn | Mus musculus | Premature transcriptional termination | Growth deficiency for paternally inherited mutation | No | (Sleutels et al., 2002) |
| Evf2 | Mus musculus | Premature transcriptional termination | None | N/A | (Bond et al., 2009) |
| BC1 | Mus musculus | Replacement of promoter and exon by PgkNeo cassette | Vulnerable to epileptic fits after auditory stimulation | No | (Zhong et al., 2009) |
| Neat1 | Mus musculus | 3 kb Promoter and 5’ deletion | None | N/A | (Nakagawa et al., 2011) |
| Tsx | Mus musculus | 2 kb Promoter and exon 1 deletion | Smaller testes and less fearful (males) | No | (Anguera et al., 2011) |
| Malat1 | Mus musculus | Deletion | None | N/A | (Eissmann et al., 2012) |
| Malat1 | Mus musculus | lacZ insertion and premature transcriptional termination | None | N/A | (Nakagawa et al., 2012) |
| Malat1 | Mus musculus | 3 kb Promoter and 5’ deletion | None | N/A | (Zhang et al., 2012) |
| Hotair | Mus musculus | Deletion | Spine and wrist malformations | No | (Li et al., 2013) |
| Hotdog and Twin of Hotdog | Mus musculus | Large (28 Mb) translocation by inversion | Loss of Hoxd expression in the cecum | N/A | (Delpretti et al., 2013) |
| Fendrr | Mus musculus | Replacement of exon 1 with transcriptional stop signal | Embryonic lethal around E13.75 | Yes (majority of embryos) | (Grote et al., 2013) |
| Fendrr | Mus musculus | Locus replacement with lacZ cassette | Perinatal lethality | No | (Sauvageau et al., 2013) |
| Peril | Mus musculus | Locus replacement with lacZ cassette | Perinatal lethality | No | (Sauvageau et al., 2013) |
| Mdgt | Mus musculus | Locus replacement with lacZ cassette | Reduced viability and reduced growth | No | (Sauvageau et al., 2013) |
| 15 other lncRNA loci | Mus musculus | Locus replacement with lacZ cassette | None | N/A | (Sauvageau et al., 2013) |





