Identification of a new stem cell population that generates Drosophila flight muscles
Figures
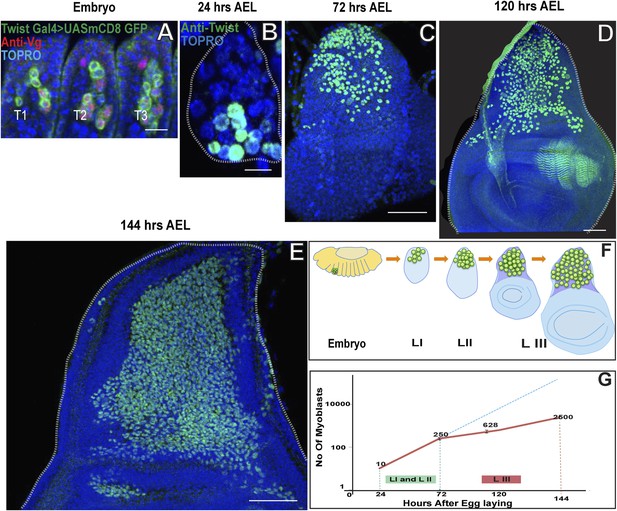
Wing disc associated AMPs proliferate during larval development to reach a population size of 2500.
(A) Stage 17 embryo (∼22 hr After Egg Laying (AEL) showing cluster of 10 AMPs in thoracic segment (T2) marked with Twist Gal4 > UAS mCD8GFP, Vg (anti-Vestigial, red) and TO-PRO3 (A nuclear stain, blue), Similar numbers of Twi positive cells are seen in each segment. n = 5 Scale bar, 10 μm. (B–E) Wing imaginal discs from early first (∼24 hr AEL) n = 5. Scale bar, 10 μm, late second instar (∼72 hr AEL) n = 10 and third instar stage (∼120 hr AEL, n = 10 and ∼144 hr AEL, n = 10) stained for Twi (anti-Twist, green) and TO-PRO3 (A nuclear stain) showing increase in the number of AMPs during the larval instars. Scale bar, 50 μm. (F) Schematic showing AMPs, marked in green color, in T2 region of stage 17 embryo and subsequently in the presumptive notum of the first instar, second instar and late third instar wing imaginal disc. (G) A sharp increase is seen in the number of AMPs in first (I) and second (II) instars (Till 72 hr AEL) (Depicted as red line). After 72 hr AEL (Early third instar) till the end of third instar (144 hr AEL), the rate of increase of the AMP population is less sharp. The dotted blue line depicts the extrapolation of the early rate of growth. The graph depicts the average number of cells and the bar represents the standard error. For first instar (24 hr) n = 5, late second instar (72 hr) n = 10, mid third instar (120 hr) n = 10 and late third instar (144 hr) n = 10.
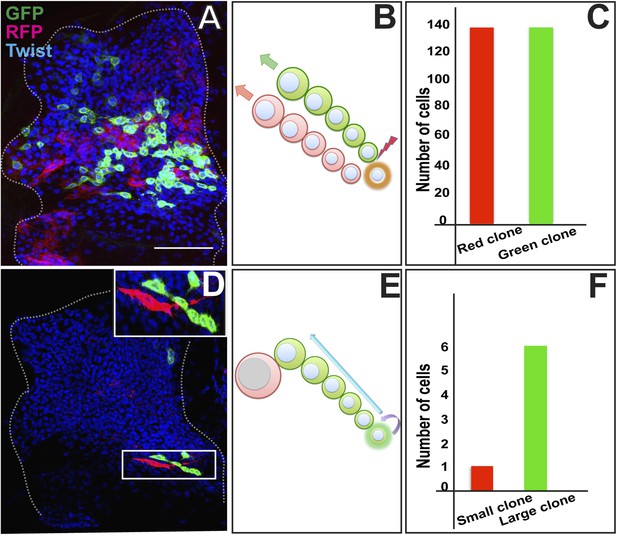
AMP proliferation involves initial symmetric and subsequent asymmetric clonal amplification.
(A) Third instar wing imaginal disc (∼144 hr AEL) showing a Twin-spot MARCM clone induced by a single15 m heat shock at 37°C in the early first instar (∼24 hr AEL). The GFP and RFP twin spots are of the same size. Anti-Twist marks all the descendants of AMP lineages. (B) Schematic depiction of clonal generation of symmetric clone from the AMP from the early first instar till late second instar stage. N = 10 early clones were examined all with a similar result showing the twin spots of the same size. (C) Quantitation of the clone showing exactly same number (136) of cells in both red and green clone of twin-spot. (D) Asymmetric clone of AMP lineage recovered from a single15 m heat shock at 37°C clonal induction in early third instar (∼75 hr AEL). Clone shows a single large red cell and six small green cells (zoomed image showed in top most right corner). Scale bar 50 μm. (E) Schematic depiction of clonal generation of asymmetric clone from the AMP in the early third instar onwards. N = 12 late clones were examined all with a similar result. (F) Quantitation of clone showing one red and six green cells from the twin-spot marking experiment.
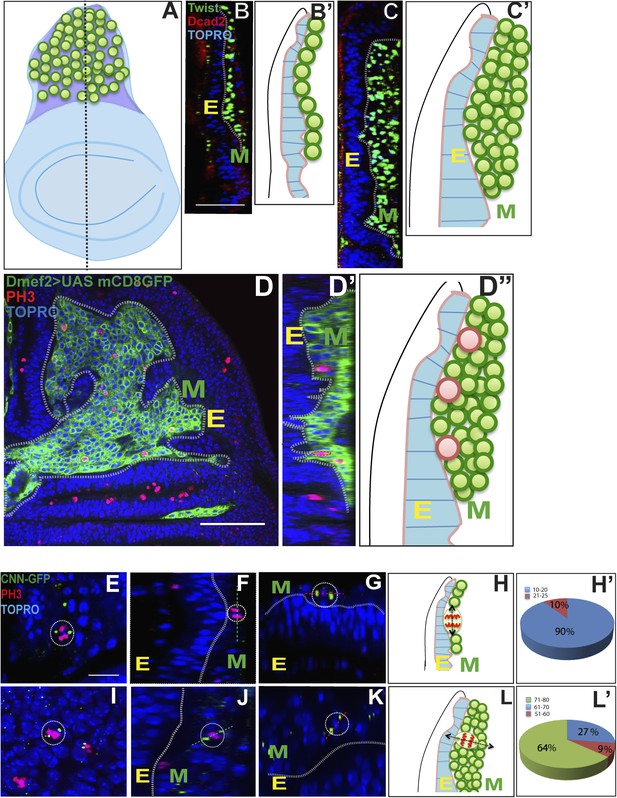
Proliferating AMPs are located in a monolayer adjacent to the wing disc epithelium.
(A–B′) Optical section of late second instar wing imaginal disc (∼72 hr AEL, schematic in A with AMPs in green), showing monostratified (single layer) arrangement of AMPs (marked in green and denoted as M for mesoderm, B) stained for Twi (anti-Twist, green), Dcad2 (anti-Dcad2, red) (Disc-epithelium [E]) and TO-PRO3 (a nuclear stain, blue). n = 10, Schematic in B′. (C–C′) Late third instar wing disc showing multistratified (3–4 layers) of AMPs in presumptive notum region. n = 20 Scale bar 50 μm. AMPs are stained for Twi (anti-Twi, green), epithelium (anti-Dcad2, red) and TOPRO (Nuclear stain, blue) (D–D″) A late third instar wing imaginal disc showing multistratified arrangement of AMP using Dmef2-Gal4 > UASmCD8GFP (anti-GFP, green) and co-stained for mitotic marker PH-3 (anti-phosphohistone, red) and TO-PRO3 (Nuclear stain, blue). C′ shows a schematic representation. D′ shows an optical section of image D showing active mitotic divisions (red dotted circles) exclusively in the layer most-proximal to the disc epithelium (marked as E). n = 20. D″ represents this schematically. (E, F and G) Late second instar wing discs (∼70 hr AEL) marked for centrosomin-GFP (CNN-GFP, a pericentrosomal marker, green) using Dmef2-Gal4, PH3 (anti-phosphohistone, red) and TO-PRO3 (Nuclear stain, blue). In each panel, E is the Epidermis and M the AMPs. (H–H′) Schematic (H) of a late second instar disc (as shown in F) showing active mitotic division (Black arrow) parallel to the disc epithelium. Pie-chart representation (H′) (Blue: 10°–20° with 0° being parallel to the epidermis. n = 6 preps. (I, J and K) Third instar wing imaginal disc (∼140 hr AEL) showing a more orthogonal orientation in AMP marked using centrosomin-GFP (CNN-GFP) (a centrosome marker, green) using Dmef2-Gal4, PH3 (anti-phosphohistone, red) and TO-PRO3 (Nuclear stain, blue). n = 10. Scale bar, 10 μm. (L–L′) A schematic representation (L) and pie-chart representation (L′) of late third instar disc showing mitotic division axis (black arrow, in L) of AMP (green) oblique to disc surface (blue). The angle of division at this stage is in the range of 50–80° as represented in L′.
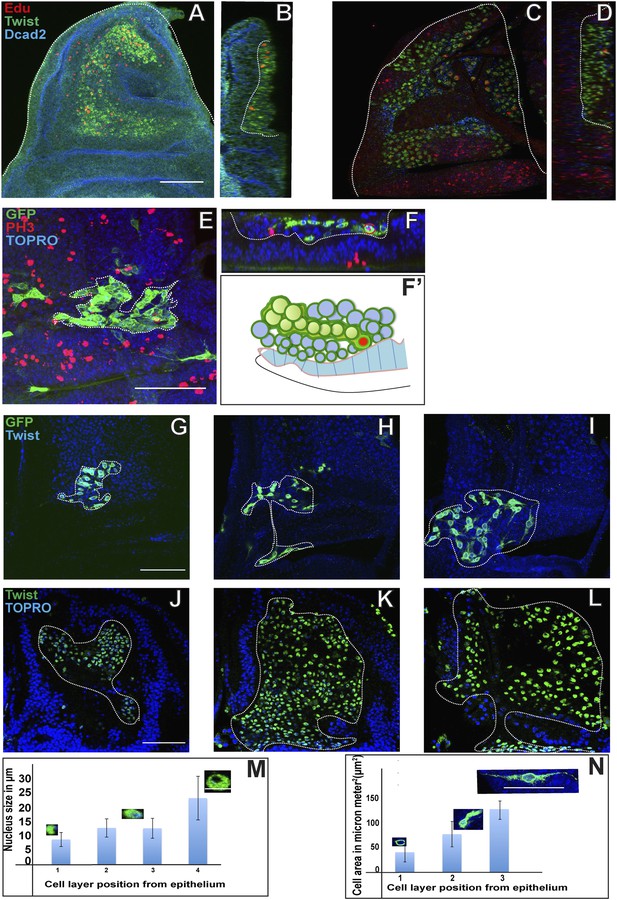
Proliferating AMPs generate clones of post mitotic myoblasts localized in distal layers.
(A) Optical section of third instar wing disc dissected after 5 hr Edu, a thymidine analogue (5-ethynyl-2′-deoxyuridine) pulse and stained for Edu (red), Twi (anti-Twist, green) and Dcad2 (anti-Dcad2, blue) revealing presence of Edu in some AMP lineage. n = 10. Scale bar, 50 μm. (B) Sagittal section of presumptive notum region of disc (shown in A) showing Edu labeling of AMP lineage only next to disc epithelium surface. (C) Third instar disc dissected after 5 hr Edu pulse and 48 hr chase in pulse free media and stained for Edu (red), Twi (anti-Twist, green) and Dcad2 (anti-Dcad2, blue) showing labeling in the most distal layer of AMP lineages. n = 12. (D) Sagittal section of C showing labeling in maximum labeling in distal layers and minimum in the layer next to epithelium. (E) A MARCM clone generated in second instar stained for GFP (anti-GFP, green) and PH3 (anti-Phosphohistone, red), TO-PRO3 (blue). The clone shows a single PH3 positive cell (shown in red bracket) in the cluster of other clonal progenies. n = 10. (F–F’) Optical section (F) and schematic (F’) of figure E, showing the presence of PH3 positive AMP (red bracket) next to the disc epithelium (dotted line). (G–I) The MARCM clone showing clonal progenies marked with GFP in the proximal most (G), middle (H) and distal most (I) layers with reference to the disc epithelium. The graph (N) shows increase in the cell size from G to J. n = 12 Scale bar 50 μm. (K–N) The optical sections of late third instar disc stained for Twi (anti-Twist, green) and TO-PRO3 (blue) and the quantitation (M) showing trend of increase nuclear size in the layers distal most in reference to epithelium. n = 10. Scale bar 50 μm.
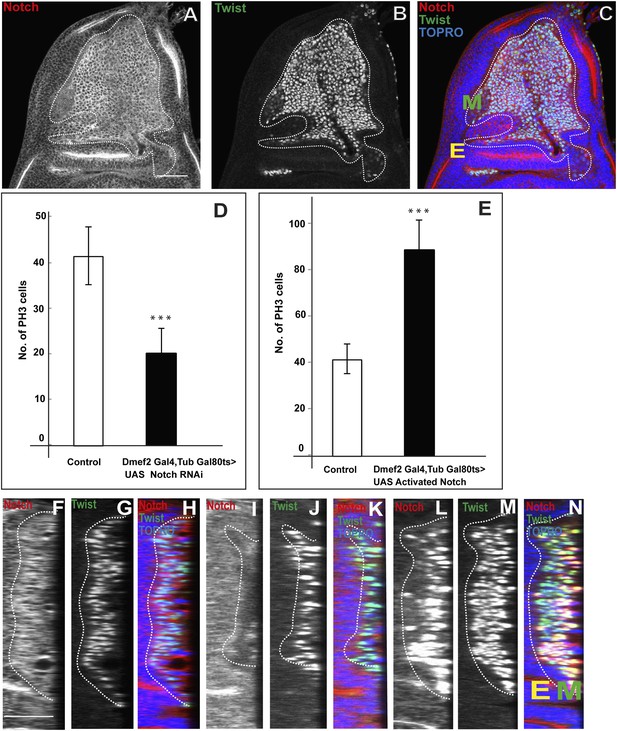
Notch signaling is required for proliferative activity of AMP lineages.
(A–C) Late third instar disc stained for Notch (anti-Notch intracellular C-terminal domain, [NICD, red]), Twi (anti-Twist, green) and TO-PRO3 (blue). In this figure, E denotes disc-epithelium and M is for mesoderm (green). (D–E) Quantification of number of PH3 positive AMPs in the Notch donwregulation using Dmef2-Gal4, TubGal80ts > UAS Notch RNAi (D) and Notch up regulation using Dmef2-Gal4, TubGal80ts > UAS NICD background (E). For both experiments Gal80 repression was relieved from early second instar till late third instar by shifting from 18°C to 29°C. All graphs are Mean ± Standard Error (Student's t test). n = 12. p-values < 0.001. (F–N) Optical section of wing discs stained for Notch (anti-Notch intracellular C-terminal domain, (NICD, red), Twi (anti-Twist, green) and TO-PRO3 (blue) in control (F–H) (Dmef2-Gal4, TubGal80ts > Canton-S), in Dmef2-Gal4, TubGal80ts > UAS Notch RNAi (I–K), Dmef2-Gal4, TubGal80ts > UAS NICD (L–N). The multistratified layered arrangement of AMP lineages is lost in Notch down regulation (J) while in Notch up regulation (M) it increases with respect to number of layers, in comparison to control (G). Gal80 repression was relieved from early second instar till late third instar by shifting from 18°C to 29°C. n = 20. Scale bar, 50 μm. E denotes disc-epithelium and M is for mesoderm (marked by anti-Twist, green).
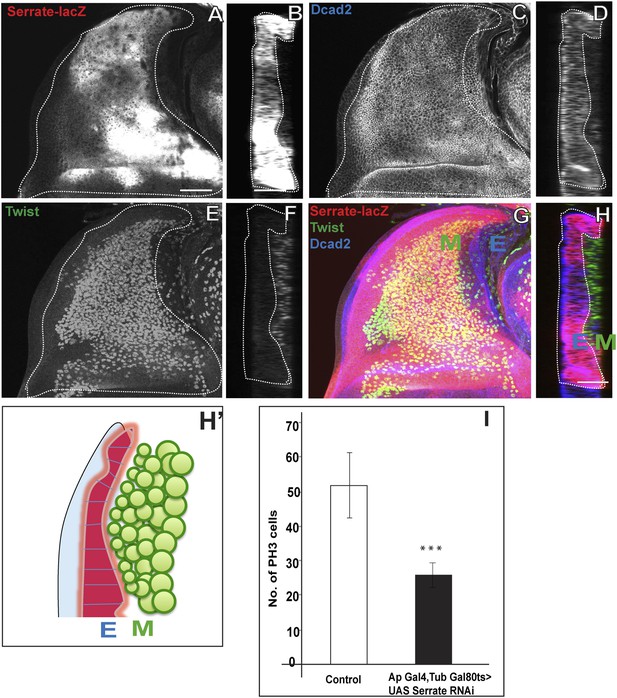
Serrate located in the wing disc epithelium is necessary for AMP proliferation.
(A) Serrate-lacZ (anti-beta Gal), a reporter for Serrate expression, visualized in disc epithelium of the late third instar. (B) Saggital section shows Serrate being expressed specifically in the disc epithelium. (C–D) Dcad2 expression (anti-Dcad2) marking the disc epithelium. (E–F) AMP lineages viewed by using Twi staining (anti-Twist). (G–H) The merge shows the expression of Serrate (red) exclusively in the disc epithelium as Dcad2 (blue) and in close proximity of first layer AMP lineages. E-wing disc epithelium, M-AMP lineages. Scale bar 50 μm. H′—Schematic depiction of expression patterns of Serrate, Dcad2 and Twist showing only one layer of AMP out of 3–4 (As shown in F) is in direct contact with Serrate producing epithelium surface. (I) Quantification of number of PH3 positive AMPs in disc epithelium specific Serrate knock down using Ap Gal4 driving UAS Serrate RNAi. Gal80 repression was relieved from early second instar till late third instar by shifting from 18°C to 29°C. All graphs are Mean ± Standard Error (Student's t test). n = 15. p-values < 0.001.
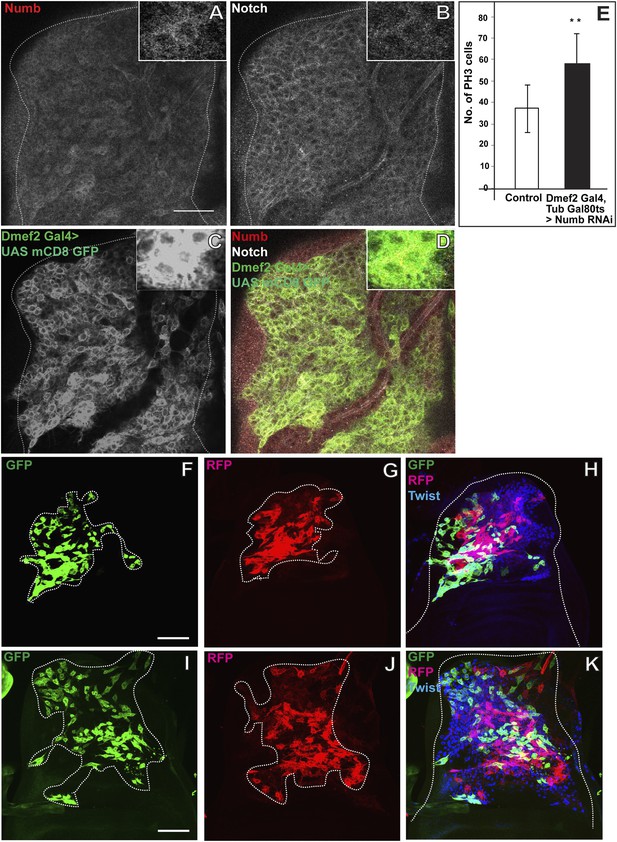
Numb expressed in the third instar stage is required for asymmetric cell divisions in AMP lineages.
(A–D) The late third instar disc stained for expression of Numb (anti-Numb, red), Notch (anti-NICD, white) and Dmef2-Gal4 > UAS mCD8GFP (anti-GFP, green). N = 6.The expression of Numb can be seen in patches (A) in contrast to Notch (B) which stains most of the AMP lineages marked by using Dmef2-Gal4 > UAS mCD8GFP (C). The merge (D) shows the expression of Notch (white) in myoblasts lineage (green) along with Numb (red). (E) Quantification of number of PH3 positive AMPs in Numb knock down (Dmef2-Gal4, TubGal80ts > UAS Numb RNAi) showing significant increase in the total proliferating AMPs. Gal80 repression was relieved from early third instar till late third instar by shifting from 18°C to 29°C. All graphs are Mean ± Standard Error (Student's t test). n = 10. p-values < 0.001. (F–K) Twinspot MARCM in Numb RNAi (F–H) and in Notch upregulation (I–K) backgrounds, generated in early third instar, showing loss of asymmetry or symmetric clones in contrast to wild type asymmetric clone as shown in Figure 2D–F. n = 5. Scale bar, 50 μm.
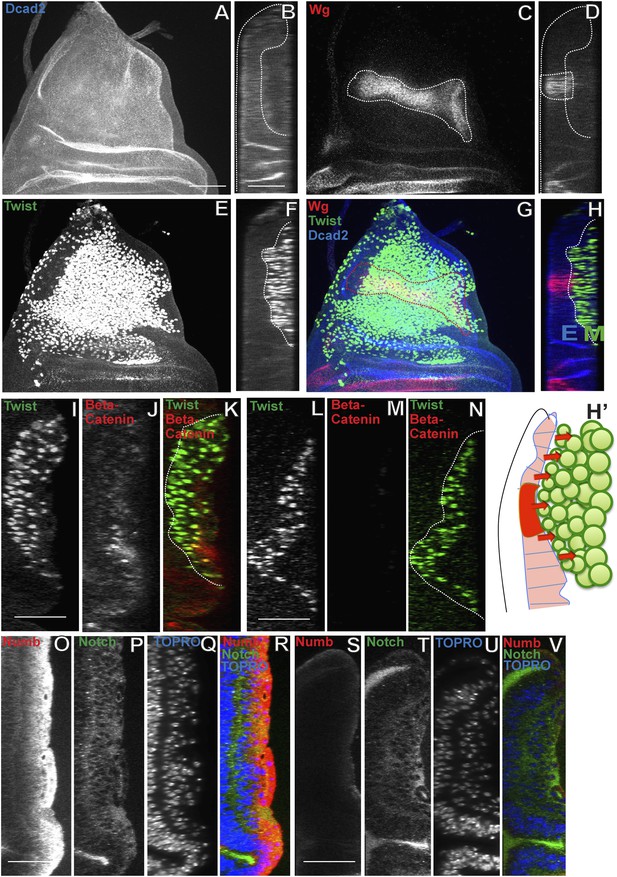
Wingless signaling from the wing disc epithelium induces Numb expression in third instar AMP lineages.
(A–H) Third instar wing disc stained for Dcad2 (anti-Dcad2, Blue), Wg (anti-Wg, red) and Twi (anti-Twist, green) demonstrating a prominent longitudinal stripe of Wg expression in disc epithelium. n = 20. (H′) Schematic of the merge (H) depicting disc epithelium, as a source of Wg production and dispersal subsequently leading to Wg signaling (red arrows) activated in all AMPs. (I–N) AMP lineages (anti-Twi, green) stained for Beta-catenin (down stream molecule of Wg pathway) (anti-Beta catenin, red) in Canton-S (I–K) and in Wg(ts)/Wg(Sp-1) alleles (loss of function alleles of Wg gene) (L–N). Loss of Beta-catenin in Wg(ts)/Wg(Sp-1) shows absence of Wg activation in AMPs which also leads to decrease in number of AMP lineages. Presence of Beta-catenin in all AMPs clearly points towards Wg action at a distance far from the disc epithelium, the source of Wg. n = 10. (O–R) The optical section of third instar wing disc showing Numb expression (anti-Numb, red) very prominent in the distal layer of AMPs, marked by Notch (anti-Notch, green) (also in Figure 5A–C). (S–V) Wg loss of function (Wg(ts)/Wg(Sp-1)) results in total disappearance of Numb in AMPs. n = 10. Scale bar, 50 μm.
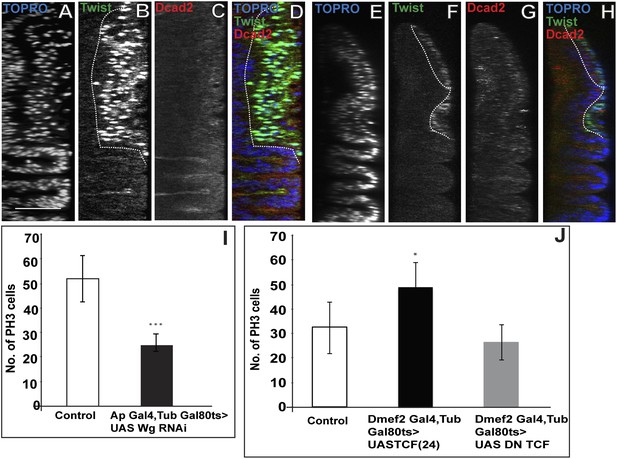
Loss of Wg results in reduction of mitotic activity and layered arrangement in AMP lineage.
(A–H) Third instar wing disc showing loss of multilayered arrangement in Wg loss of function back ground (Wg(ts)/Wg(Sp-1)). In control disc (Canton-S) the multistratified arrangement can be distinctly seen which clearly disappears in Wg loss of function background. n = 8. (I) Quantification of number of PH3 positive AMPs in epithelium specific Wg knockdown using ApGal4 > UAS Wg RNAi showing significant decreases in comparison to control (Canton-s). All graphs are Mean ± Standard Error (Student's t test). p-value < 0.001, n = 10. (J) AMP specific perturbations (using Dmef2-Gal4) of Wg pathway downstream molecules (TCF) showing changes in mitotic activity in comparison to control. The activation of Wg pathway by overexpressing activated TCF (UAS TCF 24) leads to significant increase in mitotic activity. Gal80 repression was relieved from early second instar till late third instar by shifting from 18°C to 29°C. All graphs are Mean ± Standard Error (Student's t test). n = 15.
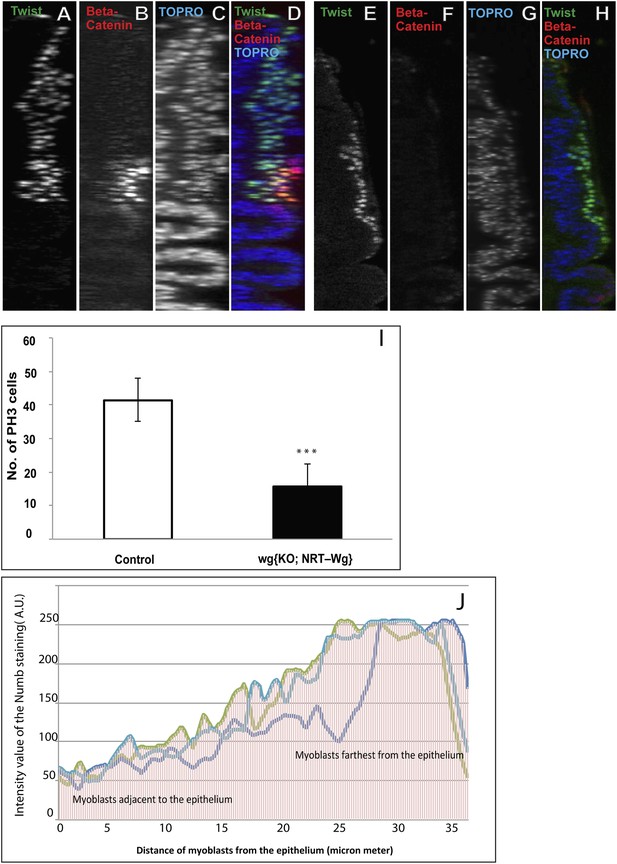
Membrane tethered Wg perturbs multistratified arrangement of AMPs.
(A–H) The third instar wing imaginal discs from wg{KO, Nrt-Wg} (tethered Wg, non-secretory form) (E–H) shows significant reduction in myoblasts layers (anti-Twist, green) and also beta-catenin (anti-Beta-catenin, red) activation in comparison to control disc (A–D). n = 8. Scale bar, 50 μm. (I) The graph showing numbers of actively dividing AMPs in wg{KO, Nrt-Wg} genotype and in control discs. The proliferation of AMPs is significantly less than control wing discs (Mean ± Standard Error (Student's t test). n = 10. p-values < 0.001). (J) Intensity of Numb expression in third instar wing discs. The expression is highest in the AMPs farthest from the epithelium. Number of discs used for analysis, n = 6.
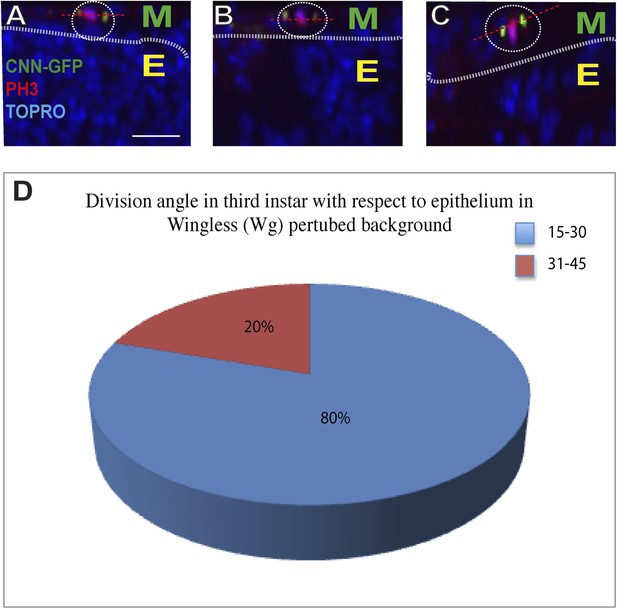
Wg down regulation alters division axis of third instar AMPs.
AMP specific perturbation (Using Dmef2-Gal4) of Wg pathway downstream molecule (DN-TCF) results in an axis of division (marked by CNN-GFP, green) parallel to disc epithelium (wing disc epithelium-E) in the third instar (A, B and C are three representative examples). n = 10. Scale bar, 10 μm. Gal80 repression was relieved from early second instar till late third instar by shifting from 18°C to 29°C. (D) A pie chart representation of the data.
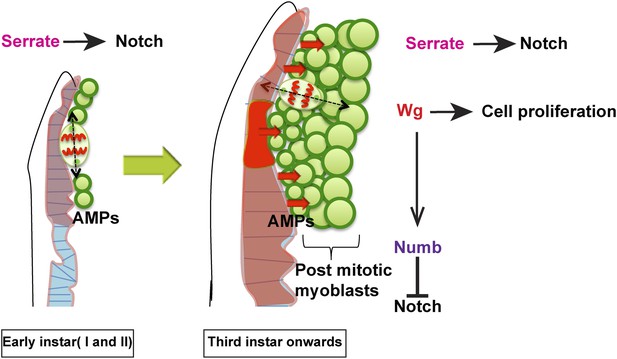
Model proposed.
In the early instars (I and II) AMPs exhibit symmetric division along epithelium and Serrate-Notch signaling plays major role at this stage. In the third instar onwards the axis of cell division in AMPs changes to orthogonal orientation and expression of Wg in disc epithelium along with Serrate-Notch signaling regulates AMP proliferation. Wg signaling potentially regulates Numb, which inhibits Notch leading to asymmetric divisions.
Videos
Showing multilayered arrangement of myoblasts (related to Figure 3).
3D reconstruction of third instar wing disc showing AMP lineage stained with anti-Twi (green) and all disc nuclei stained using TOPRO (blue) showing stacked arrangement.
MARCM clone showing variation of cell size (related to Figure 4).
3D reconstruction of AMP lineage marked by membrane tethered GFP (mCD8::GFP) revealing size differences in the myoblasts at different distances from disc epithelium. All nuclei marked by TOPRO (blue).
MARCM clone showing PH3 association with respect to layers (related to Figure 4).
Single cell in a clone (anti-GFP, green) showing active mitotic division (anti-Phospho histone 3, red).
Serrate staining in epidermis and close association with myoblasts (related to Figure 6).
Serrate- lacZ (anti-beta-gal, red) showing serrate expression in wing disc epithelium (anti-Dcad 2, blue) and only one layer of myoblasts are in contact with epithelium due to multilayered arrangement.
Wg staining in Canton-S (related to Figure 8).
Late third instar wing disc showing Wg (anti-Wg, red) expression in disc epithelium (anti-Dcad2, blue). The myoblasts forms a close association with Wg expression.





