Science Forum: Improving pandemic influenza risk assessment
Figures

Evidence for concern and actions to mitigate influenza pandemics.
Types of evidence that have been, or could be, used to justify specific preparedness or mitigation actions prior to evidence of sustained human-to-human transmission, largely based on the authors' interpretation of national and international responses to H5N1, H7N9, and H3N2v outbreaks (Epperson et al., 2013, WHO, 2011). Red indicates largely sufficient, orange partly sufficient, yellow minimally sufficient, gray insufficient. * high pathogenicity phenotype as defined by the World Organization for Animal Health (OIE)(OIE, 2013).
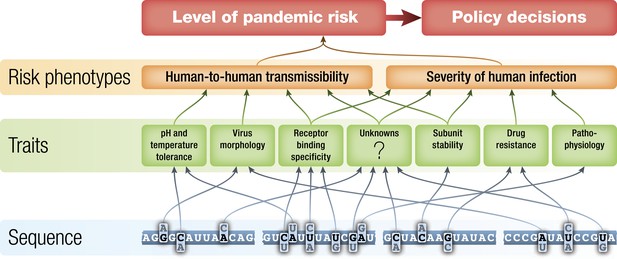
Schematic of potential relationships from virus genetic sequence to level of public health concern/pandemic risk.
Pandemic risk is a combination of the probability that a virus will cause a pandemic and the human morbidity and mortality that might result from that pandemic. Arrows represent possible relationships between levels and are not intended to summarize current knowledge.
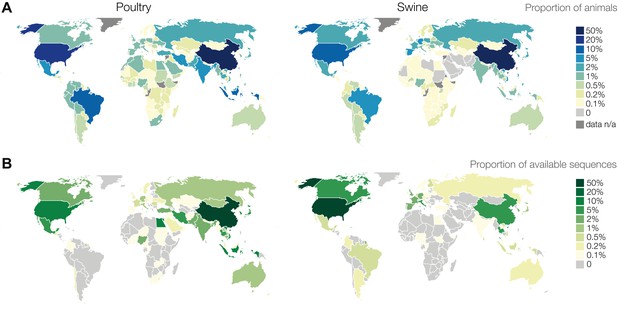
Geographic distribution of publicly available influenza virus genetic sequence data in comparison to poultry and swine populations.
(A) Proportions of worldwide animal population by country (data from the Food and Agriculture Organization of the United Nations). (B) Number of unique influenza viruses for which sequence data exists in public databases from poultry or swine by country. Numbers of influenza virus sequences are not representative of influenza virus surveillance activities. Information regarding surveillance activities is not readily available. Virological surveillance, even if robust, may result in negative findings and is not captured in these figures. Most countries do not sequence every influenza virus isolate and some countries conduct virological surveillance without sharing sequence data publicly. Sequences deposited in public databases can reflect uneven geographic distribution and interest regarding viruses of concern such as H5N1 and H9N2.





