Impaired fast-spiking interneuron function in a genetic mouse model of depression
Figures
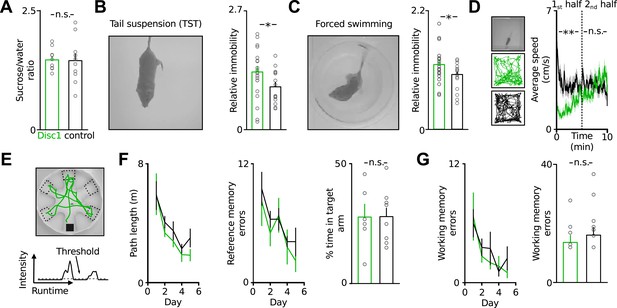
Disc1 mice show depression-related behavioural despair.
(A) Disc1 mice show similar sucrose preference as controls (146 ± 7 vs 144 ± 15% sucrose intake, n = 10 each group). (B and C) Enhanced behavioural despair of Disc1 mice in TST (37.8 ± 3.3 vs 25.8 ± 2.9%, n = 22 Disc1, 14 control mice) and forced swim test (44.1 ± 2.6 vs 37.3 ± 2.0%, n = 22, 20). (D) Unaltered locomotion of Disc1 mice. Left, examples of the path of a Disc1 and a control mouse during a 10 min exploration period (n = 19 Disc1, 18 control mice). Right, Disc1 mice move slower during the initial phase of the task but reach similar movement speeds as controls during the later phase. (E) Radial arm water maze to probe spatial reference and working memory. The animals were released from a random start arm and had to find the hidden platform in the southern arm. Green line shows the path of one representative animal during one trial. Arm entries were detected by a threshold-crossing algorithm (bottom, N = 6, 9). (F) Path length and reference memory errors plotted against the five subsequent test days. Identical shortening of the swim path length (left) and identical number of reference memory errors (entries in wrong arms; middle) of Disc1 and control mice indicates intact spatial learning. Time spent in the target arm is identical between genotypes (n = 6, 9). (G) The number of working memory errors (re-entries in previously explored arms within a trial, 14 ± 2 vs 17 ± 3) did not depend on the genotype. *p < 0.05, **p < 0.01. Data are mean ± SEM.
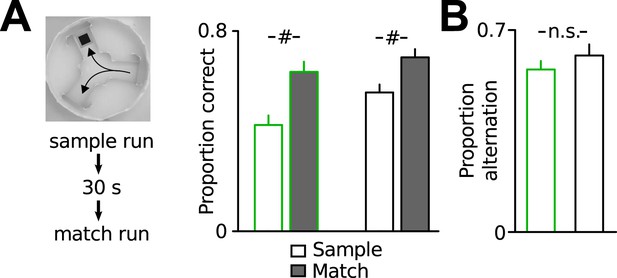
Additional assessment of working memory performance.
(A) Working memory assessment in a delayed match-to-place task in the water y-maze. In each trial, animals had to find the hidden platform in one of the target arms (sample phase). After a 30 s delay, the mice were released from the same start arm (match phase) and could either choose the correct arm or the opposite arm (working memory error). Both Disc1 and control mice performed significantly better in the match phase, indicating intact spatial working memory (n = 6, 10). (B) Both genotypes showed similar spontaneous alternation when they explored a y-maze for 10 min, confirming intact working memory (n = 8 each group). #p < 0.001. Data are mean ± SEM.
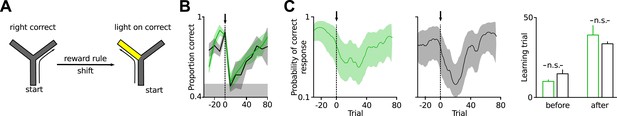
Extra-dimensional rule-shifting task in the y-maze.
(A) Mice were food-restricted and learned to forage in a y-maze. Then, they learned the first reward rule (right arm correct). In each trial, one randomly chosen arm of the two possible target arms was illuminated. Once the animals had learned the spatial rule, the reward regime was changed to ‘light on’ (extradimensional shift, n = 6, 5). (B) Normalized correct trials are plotted against trial number. Arrow at t = 0 indicates the reward rule change. Both groups of mice learned the initial spatial rule as well as the rule change. Data are binned over 10 runs. (C) Examples of individual learning curves of a Disc1 and a control mouse with 95% confidence intervals. The trial in which the lower confidence interval exceeded chance level was considered the first trial in which the animal had learned the task (learning trial). Right, identical learning trials of both rules in Disc1 and control mice. Data are mean ± SEM unless stated.
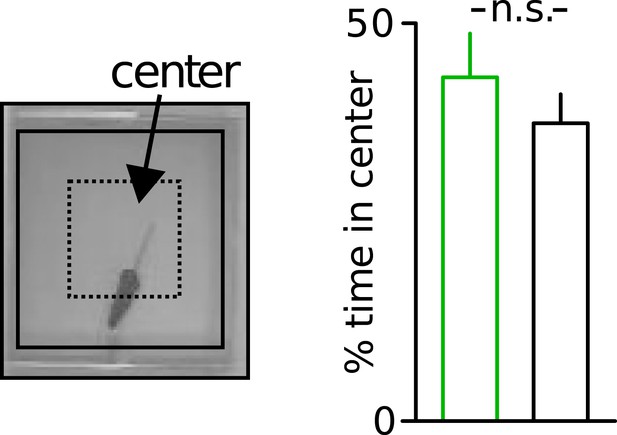
Unaltered anxiety of Disc1 mice.
Unaltered anxiety in Disc1 mice quantified from the time spent in the center of the open field (n = 19, 18). Data are mean ± SEM.
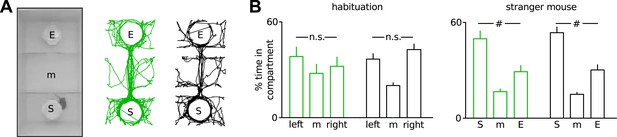
Unaltered sociability of Disc1 mice.
(A) 3-chamber social interaction task. The mice explored the 3-chamber arena containing two empty (E) wire cups in a habituation run. In a second run, a stranger mouse (S) was placed in one of the cups (n = 9 each group). Right, examples of exploration paths. (B) Disc1 and control mice spent identical time in both compartments during habituation and more time with the stranger mouse in run two, indicating absent left-right preference and intact sociability, respectively. #p < 0.001. Data are mean ± SEM.
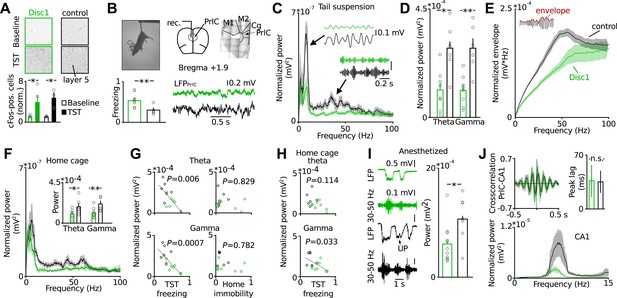
Behavioural despair of Disc1 mice correlates with impairment in theta and low-gamma oscillations in the PrlC.
(A) TST activates cFos in PrlC independent from genotype (fold increase Disc1: 3.92 ± 1.54, n = 4; control: 4.78 ± 1.07, n = 3). (B) LFP recording during TST. Enhanced freezing of Disc1 mice is preserved in the electrode-implanted cohort (52.2 ± 5.8 vs 29.4 ± 4.0, n = 8, 6). M1,2: motor cortex, Cg: cingulate cortex. (C and D) Reduced power of Disc1 mice in the theta (0.11 ± 0.03 vs 0.29 ± 0.04 mV2*10−3) and low-gamma band (0.11 ± 0.02 vs 0.29 ± 0.04 mV2*10−3, n = 8, 6). Insets: filtered traces. (E) Oscillation amplitudes over frequency. (F) Oscillatory defects are observed in the home cage (theta: 0.10 ± 0.02 vs 0.18 ± 0.04 mV2*10−3, gamma: 0.12 ± 0.02 vs 0.22 ± 0.03 mV2*10−3, n = 8, 6). (G) Theta and low-gamma power correlate with TST freezing duration (theta: r = −0.6923, p = 0.0061; low-gamma: r = −0.79, p = 0.0008) but not with home cage immobility (r = −0.029, r = −0.222). Black lines: linear fits. (H) Home cage low-gamma but not theta can predict TST freezing (gamma: r = −0.569, theta: r = −0.440). (I) Low-gamma activity in Disc1 PrlC is impaired during UP-states in anesthesia (0.6 ± 0.1 vs 1.1 ± 0.2 mV2*10−3, n = 11, 7). (J) Top, cross-correlation of LFP simultaneously recorded in hippocampus and PrlC suggests that theta oscillations are driven by hippocampus (peak lag: 36.5 ± 20.9 vs 35.3 ± 14.3 ms, n = 5, 4). Bottom, hippocampal theta power is impaired in Disc1 mice (0.87 ± 0.23 vs 4.14 ± 1.54 mV2*10−3, n = 4, 3, p = 0.01). *p < 0.05, **p < 0.01. Data are mean ± SEM, circles are individual mice.
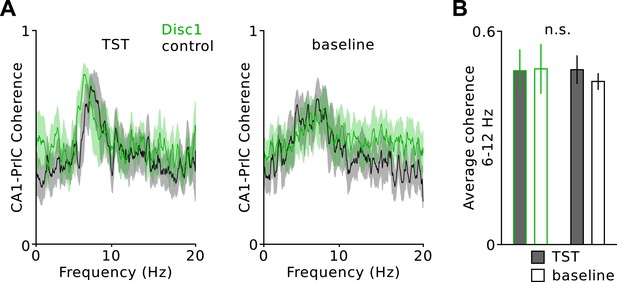
Unchanged hippocampal-prefrontal theta coherence.
(A) Average coherence during TST and baseline revealed a sharp peak in the delta frequency range in both genotypes. Solid lines are mean, shaded areas SEM. (B) There is no difference in the average coherence (6–12 Hz) between Disc1 and control mice (n = 4 Disc1, 3 control mice). Data are mean ± SEM.
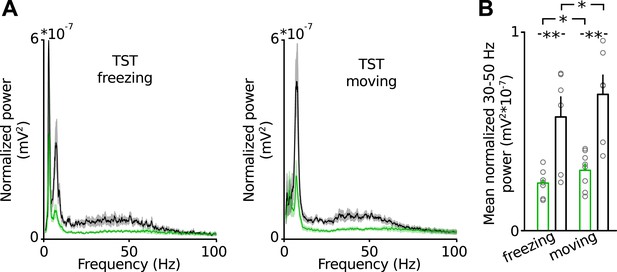
Low-gamma defect in the PrlC of Disc1 mice does not depend on the behavioral state during TST.
(A) Average power spectral density in the PrlC of Disc1 (green) and control mice (black) during freezing (left) and movement (right). (B) Summary plots of mean low-gamma power revelas significantly impaired power during both behavioral states. (n = 8 Disc1, 6 control mice). Data are mean ± SEM.
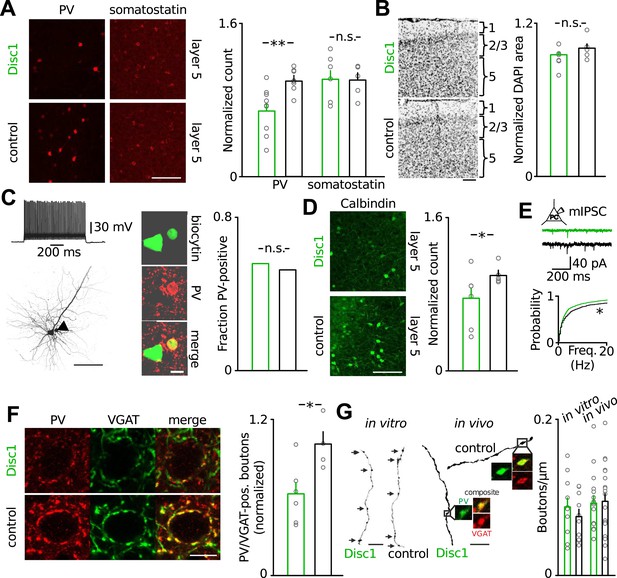
Loss of FS-INs and their output synapses in the Disc1 PrlC.
(A) Reduction of PV but not somatostatin-positive INs in the PrlC of Disc1 mice (normalized count PV: 0.69 ± 0.08 vs 1.00 ± 0.04, n = 9 Disc1, 8 control mice, p = 0.0037; somatostatin: 1.04 ± 0.10 vs 1.00 ± 0.08, n = 6, 5, p = 392). (B) Total cell density quantified from DAPI area is unchanged (normalized density: 0.93 ± 0.04 vs 1.00 ± 0.04, n = 6, 5 mice, p = 0.158). (C) FS-INs of Disc1 and control mice express PV (n = 25, 29 cells). (D) Expression of the FS-IN marker calbindin is reduced in Disc1 PrlC (0.75 ± 0.12 vs 1.00 ± 0.05, n = 6, 5 mice, p = 0.027). (E) Frequency of mIPSCs recorded in PCs was significantly reduced in the Disc1 PrlC (0.73 ± 0.14 vs 1.18 ± 0.23 Hz, n = 24, 5 cells, p = 0.025). (F) Fewer PV-VGAT-coexpressing boutons in Disc1 mice (normalized count 0.62 ± 0.09 vs 1.00 ± 0.10, n = 6, 4 mice, p = 0.021). (G) Identical bouton density of intracellularly labelled FS-INs in vitro (0.09 ± 0.01 vs 0.07 ± 0.01 µm−1, n = 12, 10, p = 0.388) and PV-positive cells in vivo (0.09 ± 0.01 vs 0.09 ± 0.01 µm−1, n = 16, 20, p = 0.868). *p < 0.05, **p < 0.01. Scale bars: A, B, C (left), D: 100 µm, E: 25 µm, C (right): 10 µm. Data are mean ± SEM, circles individual mice or cells (C and F).
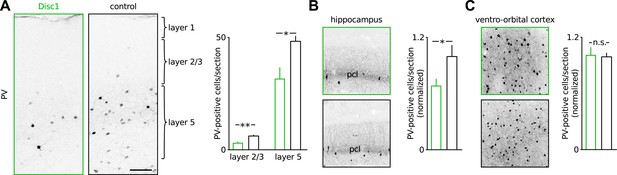
PV-positive INs are lost throughout cortical layers in the Disc1 PrlC.
(A) Immunohistochemical staining of PV in the PrlC of a Disc1 and a control mouse. Scale bar: 100 µm. Right, quantification revealed reduced PV cell counts in layers 2/3 and 5 (n = 6 Disc1, 5 control mice). (B and C) Reduction of PV cells in the hippocampus but not ventro-orbital cortex of Disc1 mice. Pcl: pyramidal cell layer. *p < 0.05, **p < 0.02. Data are mean ± SEM.
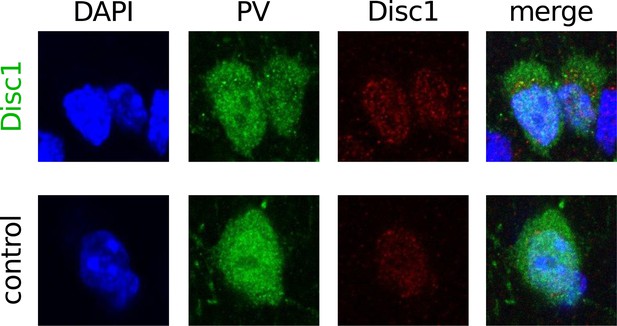
PV-positive INs express Disc 1.
Confocal image stack of DAPI, PV and Disc1 stained-sections demonstrated expression of Disc1 in PV-positive neurons in the PrlC of Disc1 and control mice. Images are representative for 35 PV/Disc1-positive cells (of 39 tested PV-expressing neurons) in Disc1 PrlC and 15 PV/Disc1-positive cells (of 15 tested) in ctrl PrlC. p = 0.832. Image size: 20 µm.
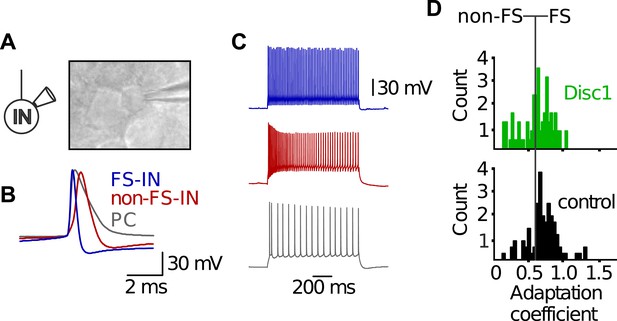
Electrophysiological characteristics of FS-INs.
(A) In vitro whole-cell patch clamp recording from a FS-IN. Infrared-differential interference contrast microscopical image reveals small and round soma shape of a FS-IN surrounded by larger PC somata. (B) FS-INs can be identified by the kinetic properties of single action potentials. Shown are superimposed examples of a single action potential of a FS-IN (blue), a non-FS-IN (red), and a morphologically identified PC (grey). FS-INs fire briefer spikes than the other cell types. (C) FS-INs respond to long-lasting somatic suprathreshold current injection (1 s) with a characteristic non-adapting, high-frequency train of action potentials. Non-FS-INs and PCs show varying degrees of accomodation and/or adaptation. (D) Adaptation coefficient defined as the first interspike interval of a train divided by the last one was used to separate FS-INs (coefficient >0.6, n = 59 Disc1, 72 control cells) from non-FS-INs (<0.6). Note, FS-INs in Disc1 and control PrlC show similar adaptation coefficients.
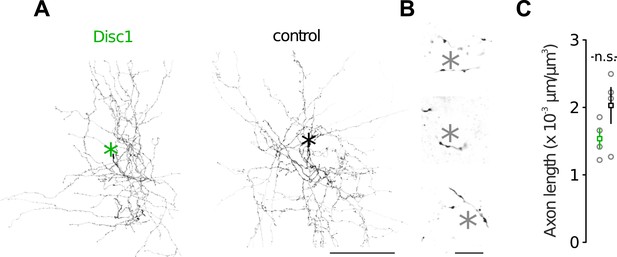
Morphological characteristics of FS-INs.
(A) Axon reconstructions of FS-INs revealed dense axonal plexus. Asterisks mark som position (not shown). (B) FS-INs form perisomatic boutons on their target cells. Asterisks indicate soma positions of putative PCs. This example is depicted from a control mouse. (C) Disc1 and control FS-INs had similar total axon lengths. Data are mean ± SEM. n = 4 cells each group.
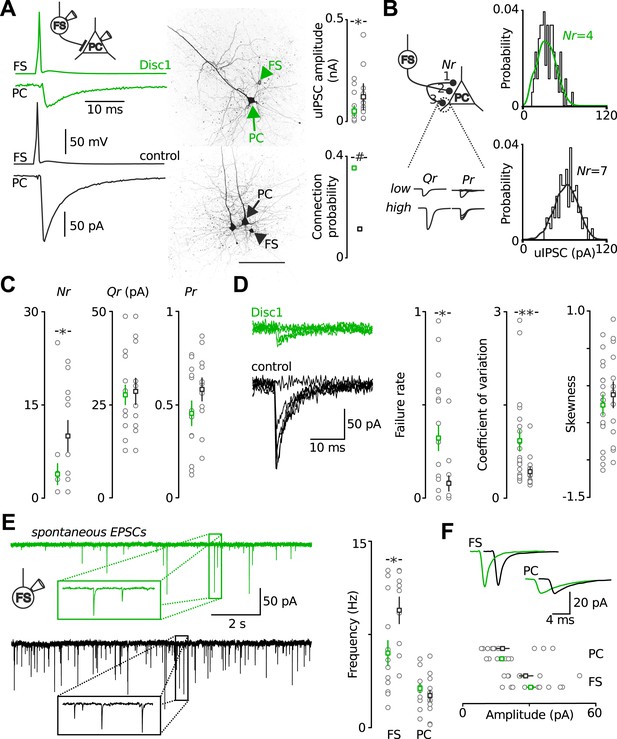
Output and input signalling of FS-INs are impaired in the Disc1 PrlC.
(A) Paired recordings of FS-INs and PCs revealed a reduction in uIPSC amplitude in the PrlC of Disc1 mice (50.8 ± 13.3 vs 121.1 ± 32.1, n = 20, 13 pairs, p = 0.012) but enhanced connection probability (35.4 vs 11.5%, n = 65, 122 simultaneous recordings, p < 0.001). Scale, 100 µm. Middle, confocal images of the pairs. (B) Amplitude distributions of uIPSCs of a Disc1 and control pair. Lines represent best fit results obtained by multiple probability compound binomial analysis. (C) Nr but not Qr or Pr are reduced at Disc1 FS-IN synapses (Nr: 4 ± 2 vs 10 ± 2, p = 0.036; Qr: 27.6 ± 2.6 vs 28.5 ± 3.5, p = 0.831; Pr: 0.45 ± 0.07 vs 0.58 ± 0.06, p = 0.340, n = 14, 11 pairs). (D) Left, superimposed single traces of a Disc1 and control pair. Failure rate, coefficient of variation, and skewness of uIPSCs support reduced Nr (0.32 ± 0.07 vs 0.08 ± 0.04, p = 0.015, 0.92 ± 0.17 vs 0.43 ± 0.07, p = 0.008, −0.262 ± 0.135 vs −0.124 ± 0.174, p = 0.503, respectively, n = 17–20, 13 pairs). (E) Reduced frequency of spEPSCs in Disc1 FS-INs (6.0 ± 1.0 vs 9.5 ± 1.1 Hz, p = 0.022, n = 15, 9) but not PCs (3.1 ± 0.4 vs 2.6 ± 0.4, p = 0.246, n = 11, 13). (F) Unchanged spEPSC amplitudes (FS-INs: 30.7 ± 2.3 vs 28.7 ± 3.4 pA, p = 0.340, n = 15, 9 cells; PCs: 17.9 ± 1.1 vs 18.4 ± 3.0 pA, p = 0.246, n = 11, 13 paris). *p < 0.05, #p < 0.001. Data are mean ± SEM, circles individual cells.
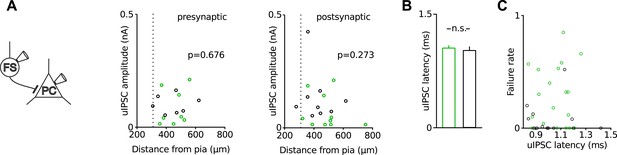
Recording depths and axonal distance between pairs of synaptically connected neurons are similar in Disc1 and control FS-IN to PC recordings.
(A) Amplitude of uIPSCs is plotted against distance of the recorded soma from the pial surface. Broken line indicates border between layer 2/3 and 5. There is no correlation between uIPSC amplitude and the anatomical depth of the presynaptic (left) or postsynaptic (right) neuron in FS-IN to PC paired recordings in acute prefrontal slices. (B) Synaptic latency was defined as the time between the maximal point of rise of the presynaptic action potential and the onset of the postsynaptic uIPSC. Paired recordings in Disc1 and control slices show similar mean synaptic latencies (Disc1: 1.0 ± 0.03 ms vs control 1.0 ± 0.05 ms, p = 0.379, n = 19 Disc1, 13 control pairs), suggesting similar axonal distance between pre- and postsynaptic partners under the assumption of similar conduction velocities of action potentials and similar transmitter release periods. (C) Failure rate did not depend on the distance (expressed as synaptic latency) between pre- and postsynaptic cells (r = −0.04, p = 0.825). Bars represent mean ± SEM, circles represent individual data points.
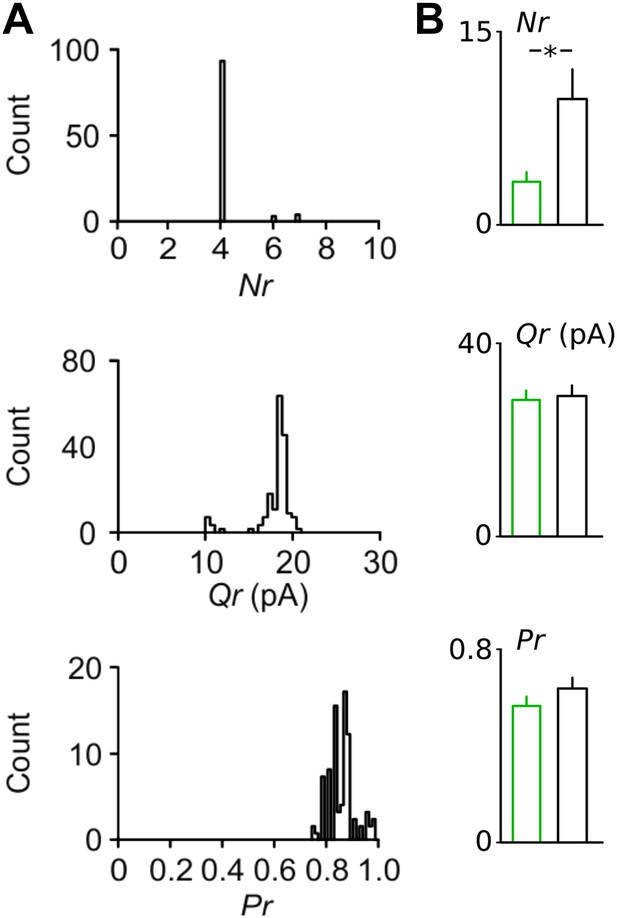
Bootstrapping analysis reveals small errors in the estimation of synaptic parameters with binomial fitting.
(A) Examples of bootstrapping of the original data from the Disc1 paired recording shown in Figure 3B. Bootstrap replications (n = 100) of the original data set were fitted in identical manner as the original data shown in Figure 4B,C. (B) Summary graphs of the synaptic parameters Nr, Pr and Qr for bootstrapping results of all pairs revealed a difference in Nr but nor Qr or Pr. Note the small errors in the estimate of all three parameters obtained. Bars represent mean ± SEM. *p < 0.05.
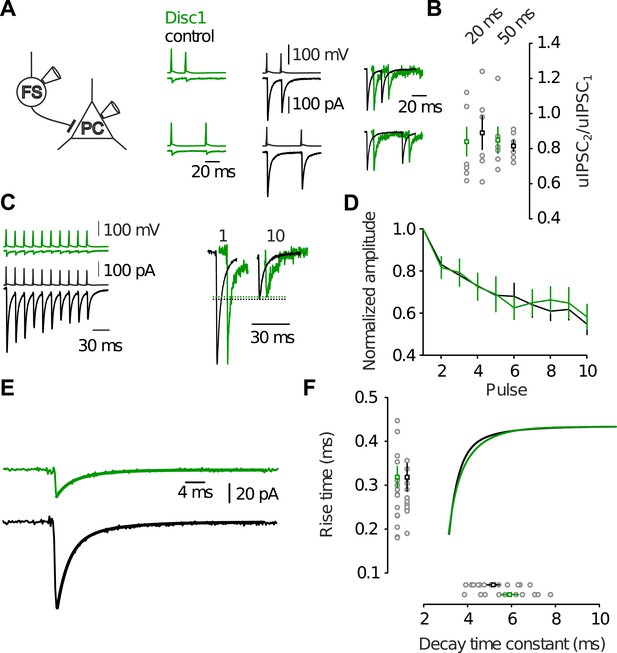
Identical dynamic and kinetic properties of uIPSCs at FS-IN output synapses.
(A) Paired-pulse modulation in FS-IN-to-PC paired recordings was was determined from dual pulse stimulation (inter-pulse interval 20 or 50 ms) in paired recordings. Two presynaptic action potentials were evoked in the FS-IN at 20 ms and 50 ms inter-pulse interval. Amplitudes of uIPSCs were measured from the preceding baseline and the paired-pulse ratio (uIPSC2/uIPSC1) was quantified. (B) Summary of paired-pulse experiments show no significant difference in short-term dynamics of uIPSCs between both genotypes (20 ms, p = 0.836, n = 7 Disc1, 6 control pairs, 50 ms, p = 0.037, n = 6, 6). (C and D) Analysis of multiple-pulse modulation of uIPSCs elicited by trains of action potentials (10 pulses, 50 Hz) in paired recordings revealed identical modulation across genotypes (10th uIPSC/1st uIPSC, p = 0.671, n = 8, 7). (E) Determination of kinetic properties of uIPSCs. The solid lines represent biexponential fits to the decay phase of the average uIPSC. (F) Neither the 20–80% rise time (p = 0.985, n = 19, 13) nor the decay time constant (p = 0.193, n = 10, 11) differed between genotypes. Inner graphs shows the amplitude-scaled fits to the decay phase shown in E. Data are mean ± SEM.
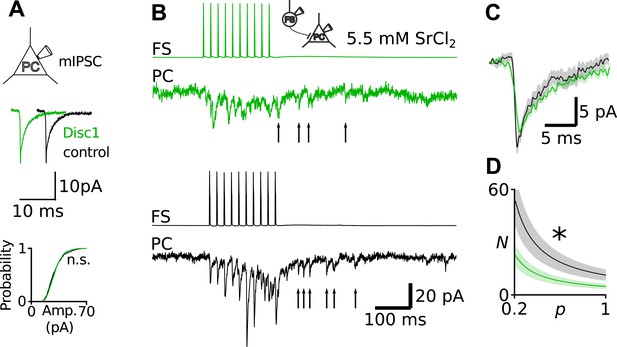
Unaltered Q at Disc1 FS-IN-to-PC synapses.
(A) Recordings of mIPSCs in PCs revealed identical amplitudes in Disc1 and control cells (N = 24, 15, p = 0.388). (B) Direct recording of quantal uIPSCs at FS-IN-to-PC synapses revealed identical Q. Quantal release was evoked in pairs in an extracellular medium containing SrCl2. Quantal uIPSCs are visible as asynchronous events following a presynaptic action potential train. (C) Superimposed average quantal uIPSCs. (D) Simulation of apparent N as a function of p. As a result of the quantification of Q shown in B and C, Q was set to 11 pA in these calculations. Independent of the value of p, N at Disc1 FS-IN output synapses is always smaller than at control pairs. *p < 0.05.
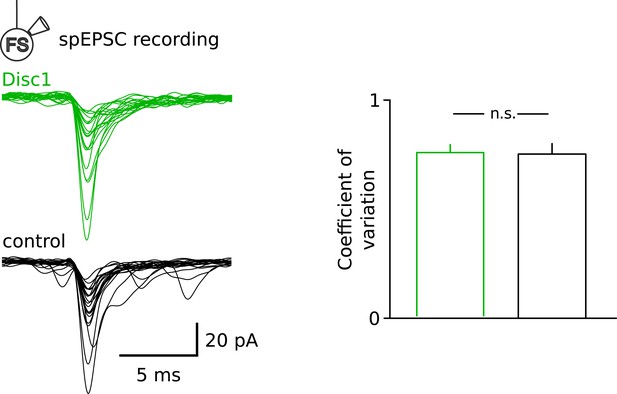
Similar fluctuation of spEPSCs at Disc1 and control FS-IN inputs.
Superimposed individual spEPSCs are shown for a representative FS-IN recording in Disc1 and control PrlC slices. Note the similar variability in the amplitude of the input spEPSCs, quantified as the coefficient of variation (p = 0.596, n = 15, 9 cells).
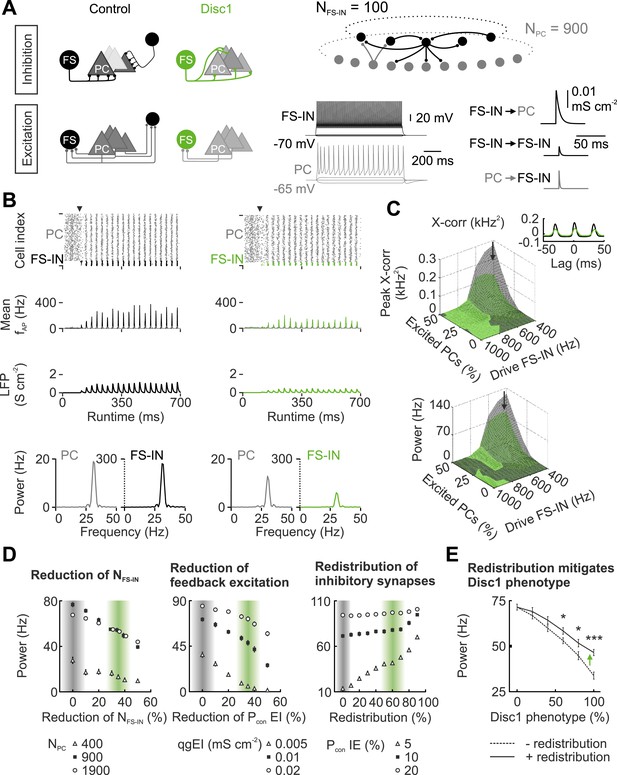
Disc1-mediated circuit changes impair low-gamma power in a network model.
(A) Schematic of network structure, cellular and synaptic properties of the control (black) and Disc1 circuit (green). (B) After synapses are enabled (arrowhead), networks synchronize in the low-gamma range. From top to bottom: Raster plots representing action potentials; binned spike frequencies; LFPs; power of firing rates. (C) Cross-correlation of PC and FS-IN activity and power under different regimes of Poisson-distributed excitatory drives. Arrows point to simulation in (B). Inset, PC-FS-IN spike cross-correlogram. (D) Effects of separate Disc1-induced circuit changes on low-gamma power. Left, effect of fewer FS-INs at different PC population sizes. Middle, reduced feedback excitation expressed as connection probability (Pcon) for different E-I quantal conductances (qg). Right, redistribution of inhibitory synapses (reduction of release sites/connection with proportional increase in Pcon I-E for networks with different mean connection probabilities). Shaded bars indicate control and Disc1 parameters. Filled squares represent the default network. (E) Low-gamma power depends on the strength of the Disc1 phenotype. Dashed line: reduced NFS-IN and Pcon EI. Continuous line: additional redistribution of inhibitory synapses. *p < 0.05, #p < 0.001.
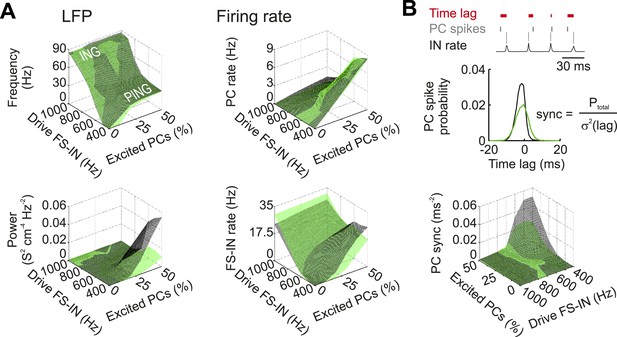
Characteristics of Disc1 and control low-gamma oscillations at different excitatory drives.
(A) Control (black) and Disc1 (green) network activity was quantified by means of the LFP (left) and the average discharge rate (right) at different excitation strengths. Excitatory drive was modelled as Poisson-distributed excitatory postsynaptic conductances. A variable fraction of the PC population (0–50%) received excitatory inputs at 0.6 kHz. In contrast, 80% of the FS-INs were excited at varying mean frequencies (0.4–1 kHz). Top left, during conditions of strong excitation of FS-INs and weak excitation of PCs, oscillation frequency was high (>60 Hz) because FS-INs were highly active and PCs were silent. This corresponds to a gamma rhythm mediated by a network of mutually connected FS-INs (interneuron gamma (ING)-mechanism). Shifting the excitation away from the FS-INs and recruiting more PCs reduced the oscillation frequency and established a gamma rhythm mediated by recurrent PC-IN connections (pyramidal-interneuron gamma (PING)-mechanism). Bottom left, highest power of gamma activity was observed in the PING regime, where most of the analysis was performed (see Figure 5C–E). (B) Top, schematic illustrating the PC sync measure as a quantification of PC spike timing relative to FS-IN activity (‘Materials and methods’). Bottom, in the PING regime (high PC drive, low FS-IN drive), PCs in the control network display much higher spike time fidelity than Disc1 PCs.
Tables
Quantitative summary of cellular and synaptic properties of Disc1 and control PrlC neurons
| Parameter | Disc1 | Control | P/N | Test | Cohen's d | |
|---|---|---|---|---|---|---|
| Sucrose preference | 1.46 ± 0.07 | 1.44 ± 0.15 | 0.901, N = 10/10 | Student's t-test | na | |
| Freezing in TST (norm.) | 1.39 ± 0.12 | 1.00 ± 0.10 | 0.015, N = 22/14 | Student's t-test | 0.82 | |
| Immobility in forced swim test (norm.) | 1.18 ± 0.07 | 1.00 ± 0.05 | 0.049, N = 22/20 | Student's t-test | 0.65 | |
| Open field total path length (m) | 14.5 ± 0.9 | 19.3 ± 0.7 | 0.0012, N = 19/18 | Student's t-test | 1.47 | |
| Open field path length (first half/second half) | 4.8 ± 0.4/8.9 ± 0.5 | 9.8 ± 0.5/9.1 ± 0.3 | 1.1*10−12, 0.682 for s half | One-way ANOVA followed by t-test | na (second half) | |
| Radial arm maze reference/working memory errors | 22 ± 5/14 ± 2 | 25 ± 2/17 ± 3 | 0.239/0.361, N = 6/9 | Mann–Whitney U test | na | |
| Proportion spontaneous alternation | 0.60 ± 0.03 | 0.65 ± 0.04 | 0.361, N = 8/8 | Student's t-test | na | |
| Extradim. spatial/shifted rule learning trial (y-maze) | 10 ± 1/39 ± 7 | 15 ± 3/34 ± 9 | 0.154/0.632, N = 8/7, 6/5 | Student's t-test | na | |
| Time in center of open field (%) | 43.5 ± 0.9 | 37.4 ± 3.5 | 0.377, N = 19/18 | Student's t-test | na | |
| 3-chamber social interaction: stranger preference | 49.4 ± 4.6 | 53.2 ± 3.3 | 0.513, N = 9/9 | Student's t-test | na | |
| TST-dependent cFos increase PrLC (norm.) | 3.92 ± 1.54 | 4.78 ± 1.01 | 0.03/0.04, N = 4/3 | Mann–Whitney U test | 1.52/3.49 | |
| Freezing in TST (%, electrode-implanted sample) | 52.2 ± 0.1 | 29.4 ± 0.1 | 0.004, N = 8/6 | Mann–Whitney U test | 1.82 | |
| TST: low-gamma power/amplitude (*10−3) | 0.11 ± 0.02/0.87 ± 0.05 | 0.29 ± 0.04/1.34 ± 0.09 | 0.003/0.002, N = 8/6 | Mann–Whitney U test | 2.33/2.77 | |
| TST: theta power/amplitude (*10−3) | 0.11 ± 0.03/0.04 ± 0.004 | 0.29 ± 0.04/0.07 ± 0.004 | 0.012/0.001, N = 8/6 | Mann–Whitney U test | 1.97/3.12 | |
| Home cage: low-gamma power (*10−3) | 0.12 ± 0.02 | 0.22 ± 0.03 | 0.009, N = 8/6 | Mann–Whitney U test | 1.62 | |
| Home cage: theta power (*10−3) | 0.1 ± 0.02 | 0.18 ± 0.04 | 0.031, N = 8/6 | Mann–Whitney U test | 1.29 | |
| Urethane anesthesia: low-gamma power (*10−3) | 0.6 ± 0.2 | 1.1 ± 0.2 | 0.024, N = 11/7 | Student's t-test | 1.27 | |
| TST: hippocampal theta power (*10−3) | 0.8 ± 0.2 | 4.1 ± 1.5 | 0.009, N = 5/4 | Mann–Whitney U test | 1.71 | |
| CA1-PrLC theta coherence TST/home cage | 0.52 ± 0.06/0.52 ± 0.07 | 0.52 ± 0.04/0.49 ± 0.02 | 0.972, N = 4/3 | One-way ANOVA | na | |
| PV-IN count PrLC all layers (normalized) | 0.69 ± 0.08 | 1.00 ± 0.04 | 0.004, N = 9/8 | Student's t-test | 1.81 | |
| PV-IN count layer 2–3/layer 5 | 3.0 ± 0.7/31.9 ± 5.3 | 6.2 ± 0.54/48.8 ± 2.52 | 0.007/0.011, N = 6/5 | Mann–Whitney U test | 2.35/1.86 | |
| Somatostatin-IN count (normalized) | 1.04 ± 0.10 | 1.00 ± 0.08 | 0.392, N = 6/5 | Mann–Whitney U test | na | |
| DAPI density PrLC (normalized) | 0.93 ± 0.04 | 1.00 ± 0.04 | 0.158, N = 6/5 | Mann–Whitney U test | na | |
| Calbindin-IN count (normalized) | 0.75 ± 0.12 | 1.00 ± 0.05 | 0.027, N = 6/5 | Mann–Whitney U test | 1.2 | |
| PV-VGAT-positive boutons PrLC (normalized) | 0.62 ± 0.09 | 1.00 ± 0.10 | 0.021, N = 6/4 | Mann–Whitney U test | 2.02 | |
| FS-IN bouton density in vitro (µm−1) | 0.086 ± 0.011 | 0.073 ± 0.009 | 0.388, N = 12/10* | Student's t-test | na | |
| FS-IN bouton density in vivo (µm−1) | 0.091 ± 0.010 | 0.093 ± 0.010 | 0.868, N = 16/20* | Student's t-test | na | |
| FS-IN axon length in vitro (mm) | 1.54 ± 0.14 | 2.02 ± 0.27 | 0.200, N = 4/4* | Mann–Whitney U test | na | |
| Amplitude (pA) | 50.8 ± 13.3 | 121.1 ± 32.1 | 0.011, N = 20/13* | Mann–Whitney U test | 0.79 | |
| rise time (ms) | 0.32 ± 0.02 | 0.32 ± 0.03 | 0.985, N = 19/13* | Mann–Whitney U test | na | |
| decay time constant (ms) | 5.92 ± 0.40 | 5.28 ± 0.28 | 0.193, N = 10/11* | Mann–Whitney U test | na | |
| onset latency (ms) | 1.01 ± 0.03 | 1.02 ± 0.05 | 0.378, N = 19/13* | Mann–Whitney U test | na | |
| FS-IN-to-PC uIPSC | failure rate | 0.32 ± 0.07 | 0.08 ± 0.04 | 0.015, N = 20/13* | Mann–Whitney U test | 1.06 |
| coefficient of variation | 0.924 ± 0.165 | 0.426 ± 0.072 | 0.008, N = 17/13* | Mann–Whitney U test | 0.92 | |
| skewness | −0.262 ± 0.135 | −0.124 ± 0.174 | 0.503, N = 17/13* | Mann–Whitney U test | na | |
| paired-pulse ratio 20/50 ms | 0.84 ± 0.08/0.85 ± 0.08 | 0.89 ± 0.09/0.82 ± 0.03 | 0.836/0.937, N = 7/6, 6/6 | Mann–Whitney U test | na | |
| multiple-pulse 50 Hz 10th | 0.58 ± 0.06 | 0.54 ± 0.05 | 0.671, N = 8/7* | Student's t-test | na | |
| Connection probability (%) | 34.6 | 12.8 | 5.3*10–29, N = 78/148* | Chi2 test | na | |
| Nr | 4 ± 2 | 10 ± 2 | 0.036, N = 14/11* | Mann–Whitney U test | 0.87 | |
| Binomial fitting | Qr | 27.6 ± 2.6 | 28.5 ± 3.5 | 0.831, N = 14/11* | Student's t-test | na |
| Pr | 0.45 ± 0.07 | 0.58 ± 0.06 | 0.178, N = 14/11* | Student's t-test | na | |
| amplitude (pA) | 30.7 ± 2.3 | 28.7 ± 3.4 | 0.34, N = 15/9* | Mann–Whitney U test | na | |
| spEPSCs on FS-INs | frequency (Hz) | 6.0 ± 1.0 | 9.5 ± 1.1 | 0.022, N = 15/9* | Student's t-test | 1.1 |
| coefficient of variation | 0.75 ± 0.04 | 0.75 ± 0.05 | 0.596, N = 15/9* | Student's t-test | na | |
| amplitude (pA) | 17.9 ± 1.1 | 18.4 ± 3.0 | 0.202, N = 11/13* | Mann–Whitney U test | na | |
| spEPSC on PCs | ||||||
| frequency (Hz) | 3.1 ± 0.4 | 2.6 ± 0.4 | 0.246, N = 11/13* | Mann–Whitney U test | na | |
| amplitude (pA) | 20.1 ± 1.1 | 21.9 ± 1.9 | 0.388, N = 24/15* | Student's t-test | na | |
| mIPSC on PCs | ||||||
| frequency (Hz) | 0.74 ± 0.14 | 1.18 ± 0.23 | 0.025, N = 24/15* | Mann–Whitney U test | 0.57 |
-
N indicates number of animals except for: *N indicates number of cells, #N indicates number of axons.
Summary of default paramters defining intrinsic and synaptic properties in network models
| Synapse | Pcon (%) | Pcon Disc1 (%) | Sdcon (IN–IN spacings) | gq (mS cm−2) | Nr | Nr Disc1 | τrise (ms) | τdecay (ms) | Esyn (mV) | ggap (nS) |
|---|---|---|---|---|---|---|---|---|---|---|
| IN-IN | 20 | 50 | 30 | 0.005 | 10 | 4 | 0.15 | 2 | −60 | 0.01 |
| IN-PC | 10 | 25 | 30 | 0.02 | 10 | 4 | 0.2 | 5.5 | −60 | – |
| PC-IN | 10 | 6.5 | 20 | 0.01 | 5 | 5 | 0.1 | 1 | 0 | – |
| exc. Drive | 0.05 | 1 | 1 | 0.1 | 2 | 0 | – |
-
Abbreviations: Pcon, probability of synaptic connections; SDcon, standard deviation in connection probability expressed as cell-to-cell distances; qg, quantal conductance; Nr, number of release sites; τrise and τdecay, rise and decay time constant; Esyn, synaptic equilibrium potential; ggap, electrical coupling.
Additional files
-
Source code 1
Custom-made analysis tools are contained in the source code file analysiscodes.txt.
- https://doi.org/10.7554/eLife.04979.026





