The equatorial position of the metaphase plate ensures symmetric cell divisions
Figures
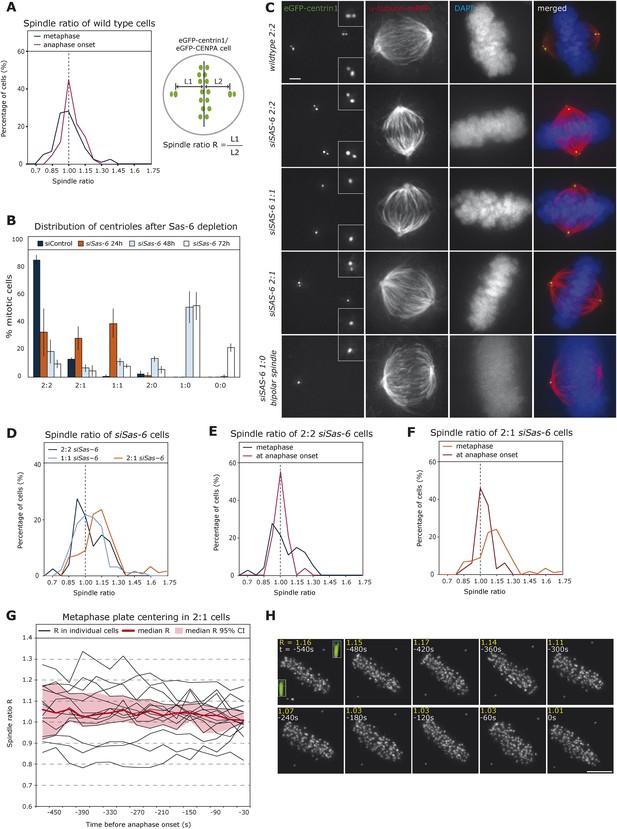
Cells center the position of the metaphase plate before anaphase onset.
(A) Distribution of spindle ratio R in metaphase cells in wild-type eGFP-centrin1/CENPA HeLa cells during metaphase in general (black curve) or 30 s before anaphase onset (red curve). The spindle ratio R was calculated by dividing the half-spindle length L1 associated with the grandmother centriole (brightest eGFP-centrin1) by the other half-spindle length L2 (for all cell numbers in all experiments see Table 1). (B) Depletion of Sas-6 but not control depletion leads to the gradual loss of centrioles after 24, 48, or 72 hr, resulting in a mixed population of cells with different number of centrioles as indicated. Centrioles were visualized based on images of eGFP-centrin1/mRFP-α-tubulin cells as shown in C (n = 50 cells per experiment, in 3 experiments, error bars indicate s.e.m.). (C) Immunofluorescence images of eGFP-centrin1 (green)/mRFP-α-tubulin (red) cells stained with DAPI (blue) with different centriole configurations as indicated. Scale bar indicates 2 μm. (D) Distribution of spindle ratio R in metaphase cells in siSas-6-transfected eGFP-centrin1/CENPA cells with different centriole configurations (2:2, 2:1, or 1:1). The spindle ratio was calculated by dividing the half-spindle length L1 associated with the grandmother centriole (2:2 and 1:1 cells) or the half-spindle length associated with 2 centrioles (2:1 cells) by the other half-spindle length L2. The spindle ratio of 2:1 cells was significantly different from the ratios seen in 2:2 or 1:1 cells (T-test with Welch's correction, 2:1 > 2:2, p = 0.018). (E, F) Distribution of spindle ratio R in Sas-6-depleted eGFP-centrin1/CENPA 2:2 (E) or 2:1 (F) cells in metaphase and before anaphase onset. 2:1 cells have a significantly more asymmetric plate position in metaphase when compared to cells just before anaphase (Mann–Whitney U test, p = 7.16 × 10−7). (G) Plot of half-spindle ratio R over time in 15 individual eGFP-centrin1/CENPA 2:1 cells that entered anaphase during live-cell imaging (black lines). The timeline was synchronized to anaphase onset (t = 0); the red curve indicates the median of R, and red surface the 95% confidence interval. Note how median R approaches 1 over time and how its variability decreases. (H) Time-lapse images of a eGFP-centrin1/CENPA 2:1 cell as analyzed in G. Half-spindle ratio R and time before anaphase are indicated for each frame. Number of centrioles was determined in IMARIS in 3D (see 3D-insets in green), * denotes the pole with 1 centriole.
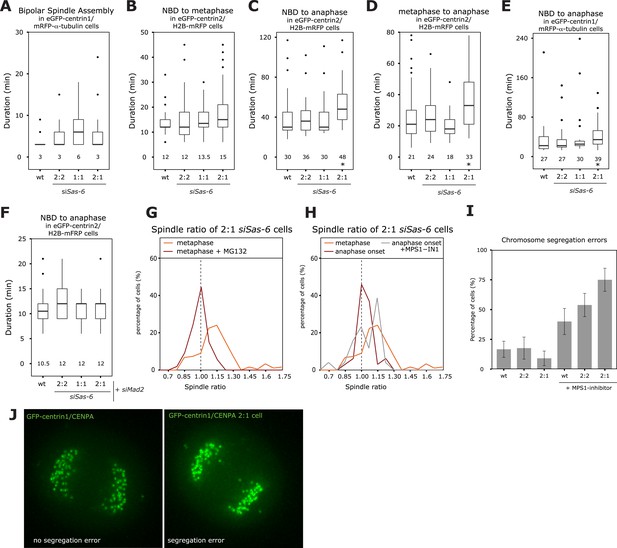
The SAC delays anaphase in cells with asymmetric spindles allowing the centering of the metaphase plate.
(A) Boxplots of the spindle assembly time (NBD until bipolar spindle formation) in wild-type and Sas-6-depleted 2:2, 1:1, or 2:1 eGFP-centrin1/mRFP−α-tubulin cells. Numbers indicate the median value, n = 49–59 cells in 2–6 experiments. (B–D) Boxplots for the time between NBD and metaphase (B); the time between NBD and anaphase onset (C); and the time between metaphase and anaphase (D) in wild-type and Sas-6-depleted 2:2, 1:1, or 2:1 eGFP1-centrin2/H2B-mRFP cells. * indicates statistically significant difference in C (Mann–Whitney U test, p = 0.003), and D (Mann–Whitney U test, p = 0.015), n = 36–100 cells in 6–13 experiments. (E) Boxplot for the time between NBD and anaphase B in wild-type and Sas-6-depleted 2:2, 1:1, or 2:1 eGFP1-centrin1/ mRFP−α-tubulin cells. * indicates statistically significant difference (Mann–Whitney U test, p = 0.003). (F) Boxplots for the time between NBD and anaphase onset in Mad2-depleted or Mad-2/Sas-6-depleted 2:2, 1:1 or 2:1 eGFP1-centrin2/H2B-mRFP cells. 2:1 cells are not delayed (Mann–Whitney U test, p = 0.836). (G) Distribution of spindle ratios R in 2:1 eGFP-centrin1/CENPA cells treated with DMSO or MG132. For cell numbers see Table 1. (H) Distribution of spindle ratio R in Sas-6-depleted eGFP-centrin1/CENPA 2:1 cells in metaphase, at anaphase onset, or at anaphase onset when treated in metaphase with the Mps1 inhibitor MPS1-IN-1. Values for metaphase and anaphase without Mps1-IN treatment were taken from Figure 1E for comparison. MPS1-IN treated 2:1 anaphase cells are significantly more asymmetric (Mann–Whitney U test, p = 0.032). (I) Quantification of chromosome segregation errors in eGFP-centrin1/CENPA cells under the indicated conditions (n = 17–30 cells; N = 4–13 experiments). Error bars indicate s.e.m. (J) Illustrative live-cell imaging stills of eGFP-centrin1/CENPA cells in anaphase with (right panel) or without (left panel) chromosome segregation errors. SAC, spindle assembly checkpoint.
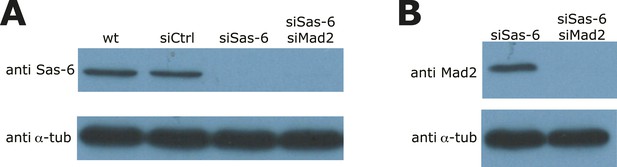
Validation of Sas-6 and Mad2 co-depletion.
(A, B) Immunoblots of wild-type, siControl-, siSas-6, or siSas-6/siMad2 treated eGFP1-centrin2/H2B-mRFP HeLa cells probed with (A) Sas-6 and α-tubulin antibodies or (B) with Mad2 and α-tubulin antibodies.
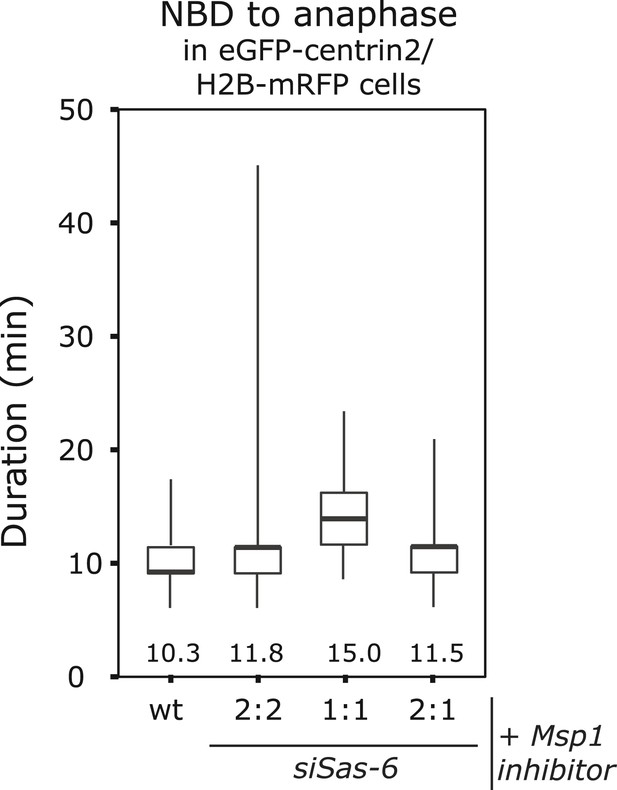
Mps1 inhibition suppresses the anaphase delay in 2:1 cells.
Boxplots for the time between NBD and anaphase onset in wild-type, siSas-6-treated 2:2, 1:1, or 2:1 eGFP1-centrin2/H2B-mRFP cells treated with an Mps1 inhibitor. Numbers indicate average anaphase times. Note that 2:1 cells are not delayed when compared to 2:2 cells (Mann–Whitney U test, p = 0.9673).
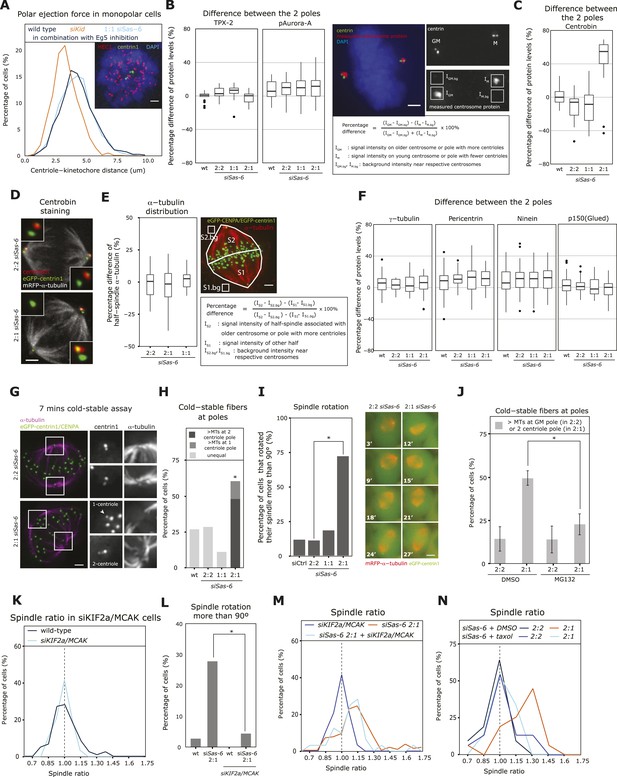
2:1 cells have half-spindles with different microtubule stability and fail to mature kinetochore–microtubule attachments.
(A) eGFP-centrin1 cells (green) were treated with monastrol and stained for HEC1 (red; right panel) to calculate the distance between kinetochores and the closest centriole. The left panel shows the distribution of centriole-HEC1 distances from n =16−28 cells, >1000 kinetochores. Scale bar indicates 2 μm. (B, C) The difference in centrosomal levels of TPX-2 and phospho-Aurora-A (B), and centrobin between each spindle pole was quantified in HeLa eGFP-centrin1 by immunofluorescence using the indicated formula, and plotted as boxplots for each centriole configuration; n = 12–63 cells. Scale bar indicates 2 μm. (D) Representative image of wild-type and siSas-6-treated 2:1 eGFP-centrin1 (green)/mRFP−α-tubulin (white) cells stained with anti-centrobin sera (red). Insets show magnified centrioles. Scale bar indicates 2 μm. (E) Quantification of the difference in α-tubulin (red signal) levels between the two half-spindles, according to the formula shown in the box. Results were plotted in the left panel using a boxplot. n = 19–25 cells, N = 2 experiments. Scale bar indicates 2 μm. (F) The difference in centrosomal levels of γ-tubulin, pericentrin, ninein and p150glued between each spindle pole was quantified as in (B) and plotted as boxplots for each centriole configuration; n = 18–68 cells. (G) Immunofluorescence images of 2:2 and 2:1 siSas-6 eGFP-centrin1/CENPA (green) cells treated for 7 min with ice-cold medium and stained with anti-α-tubulin sera (magenta). Subsetted images are maximum intensity projections of 10 stacks (z = 0.2 μm) around centrioles. Scale bar indicates 2 μm. (H) Quantification of kinetochore–microtubule minus-end stability at poles. Bar graph indicates percentage of cells that have asymmetric levels of kinetochore–microtubule minus-ends at the poles after cold-treatment; n = 26–49 cells; * indicates that within the 2:1 cell population the minus-end stability was significantly higher at the pole with 2 centriole (p = 0.00082 exact binomial test). (I) Quantification of spindle rotation in control- and Sas-6-depleted 2:2, 2:1, or 1:1 eGFP-centrin1 (green)/mRFP−α-tubulin (red) cells based on time-lapse images as shown in the right panels. Times indicate minutes after NBD. Scale bar indicates 5 μm. A spindle was counted as rotating if it had turned by more than 90° in X/Y. n = 32–122 cells in 2–6 experiments. * indicates significant difference; Fisher's exact test p = 8.39e-09. (J) Quantification of kinetochore–microtubule stability at poles. Bar graph indicates percentage of cells that have more stable kinetochore–microtubule minus-ends either at the pole with the grandmother centriole (2:2) cells or at the 2-centriole pole (2:1 cells); n = 20–40 cells in N = 3 independent experiments; * indicates that the MG132 treatment significantly reduced the percentage of cells with more stable minus-ends at the 2-centriole pole (p = 0.0275 in unpaired t-test). (K) Distribution of spindle ratios R in wild-type and siKIF2a/MCAK-treated HeLa eGFP-centrin1/CENPA cells in metaphase. For cell number see Table 1. (L) Quantification of spindle rotation in wild type, Sas-6-depleted 2:1, KIF2a/MCAK-depleted, or KIF2a/MCAK/Sas-6-depleted 2:1 eGFP-centrin2/H2B-mRFP cells, n = 18–37 cells in 1–3 experiments. * indicates significant difference; p = 0.024 in Fisher's exact test. See also Videos 1–3. (M) Distribution of spindle ratios R in Sas-6-depleted 2:1 and KIF2a/MCAK/Sas-6-depleted 2:1 eGFP-centrin1/CENPA cells in metaphase. For cell numbers see Table 1. (N) Distribution of spindle ratios R in metaphase in Sas-6-depleted 2:2 and 2:1 cells treated either with DMSO or 10 nM taxol. For cell numbers see Table 1.
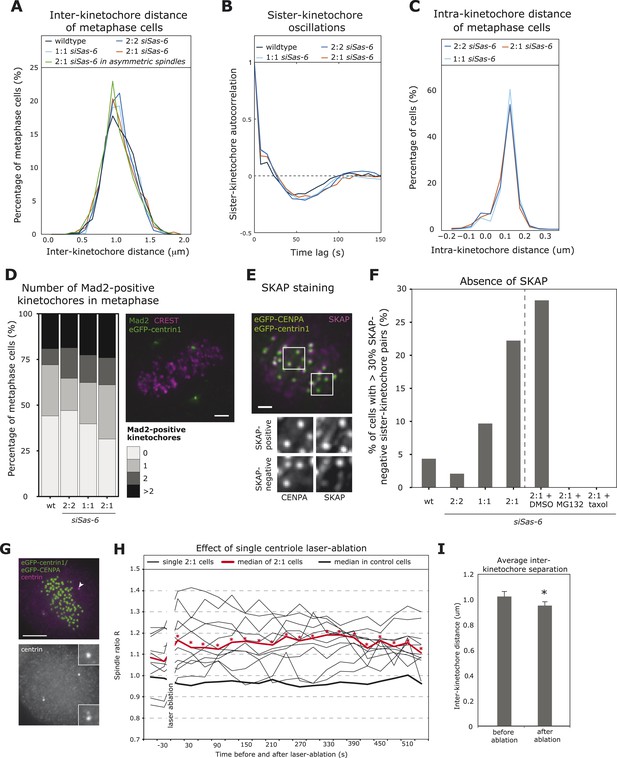
2:1 cells have immature kinetochore–microtubule attachments.
(A, B) Analysis of inter-kinetochore distances and sister-kinetochore oscillations in wild-type, Sas-6-depleted 2:2, 1:1, 2:1, or the subset of 2:1 eGFP1-centrin1/CENPA metaphase cells with an asymmetric plate position based on our in-house kinetochore tracking assay (Jaqaman et al., 2010), n = 620–889 kinetochores in 36–48 cells. The distribution of inter-kinetochore distances (CENPA to CENPA distance) is shown in A (no significant difference, t-test, p = 0.99), and the autocorrelation of the sister-kinetochore movements in B. The first minima of the autocorrelation curve indicate the half-period of the chromosome oscillations, and their depth the regularity of the oscillations. (C) Distribution of intra-kinetochore distances in wild-type and Sas-6-depleted 2:2, 1:1, or 2:1 eGFP1-centrin1/CENPA metaphase cells. Cells were stained with antibodies against the N-terminus of HEC1. Using the tracking assay, we determined for each sister-kinetochore pair the CENPA–CENPA and the HEC1-HEC1 distances, and calculated the CENPA-HEC1 distances by halving the difference, n = 701–790 kinetochores in 26–30 cells in 3 experiments (no significant difference, Mann–Whitney test, 2:1 vs 2:2, p = 0.203). (D) Quantification of Mad2-positive kinetochores in wild-type or Sas-6-depleted 2:2, 1:1, or 2:1 cells. eGFP-centrin1 (green) metaphase cells were stained with anti-Mad2 (green), and CREST sera (magenta; left panel) and the number of Mad2-positive kinetochores quantified in the right panel (n = 50–102 cells in 2 (wt) or 8 (siSas-6) experiments; no significant difference was found; Fisher's exact test, p = 0.17). Scale bar indicates 2 μm. (E, F) Quantification of SKAP-negative kinetochores in wild-type, siSas-6-depleted 2:2, 2:1, 2:1 + DMSO, 2:1, 2:1 + MG132, and 2:1 + taxol-treated eGFP-centrin1/CENPA cells. Cells were stained with antibodies against the kinetochore protein SKAP (magenta), as shown in E (maximum-intensity projection of 8 stacks [z = 0.3 μm]). Using the eGFP-CENPA (green) signal, we quantified the number of sister-kinetochore pairs with at least one SKAP-negative kinetochore (as shown in inlets). Quantification in F shows the percentage of cells where more than 30% of the sister-pairs were SKAP-negative. Fisher's exact test for 2:1 > 2:2, p = 0.0013. n = 46–72 cells in N = 4–7 experiments. Scale bar indicates 2 μm. (G) Example of an eGFP-centrin1/CENPA (green) cell in which a single daughter centriole was ablated (white arrow indicates the location of the laser pulse). Cells were fixed and stained with anti-centrin sera (magenta) to confirm the loss of a centriole, as opposed to the mere bleaching of eGFP-centrin1. Scale bar indicates 5 μm. (H) Plot of half-spindle ratio R over time in 11 single eGFP-centrin1/CENPA cells in which a single centriole was ablated. The time point of laser ablation is t = 0. The thick red curve indicates the median of R of laser ablated 2:1 cells (* denotes when median R is asymmetric [p < 0.01]), the thick black curve indicates the median R distribution of 8 control-ablated cells. (I) Average inter-kinetochore distances in eGFP-centrin1/CENPA cell before or after a single daughter centriole was ablated as determined by the kinetochore tracking assay. Error bars indicate s.e.m. n = 11 cells, 2 time points before and after ablation and on average 20 kinetochores per cell. * denotes a statistically significant difference (p = 0.001 in two-tailed paired t-test).
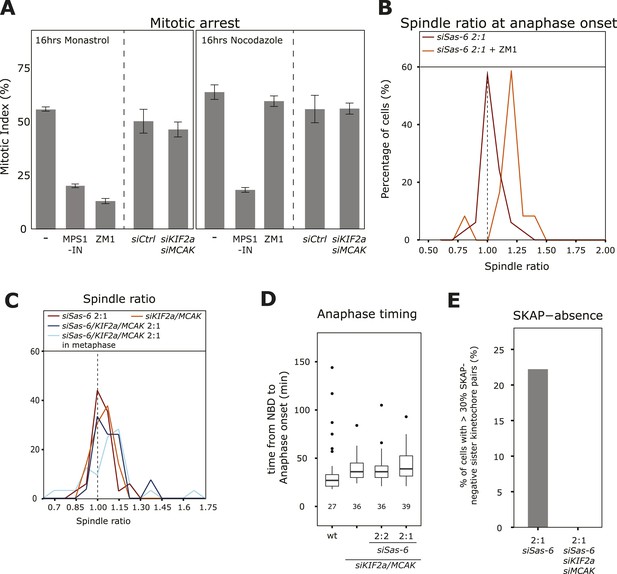
Depleting KIF2a and MCAK overcomes the SAC response in 2:1 cells.
(A) Mitotic index of untreated, ZM-1-treated, MPS1-IN-treated, control-depleted, or KIF2a/MCAK-depleted cells treated for 16 hr with nocodazole (unattached kinetochores) or monastrol (lack of tension), n ≥ 400 cells in 3–4 experiments, error bars indicate s.e.m. * ZM1 and MPS1-IN overcome a monastrol arrest (t-test p < 0.0001), and MPS1-IN overcomes a nocodazole arrest (t-test p = 0.0044). (B) Distribution of spindle ratio R in Sas-6-depleted eGFP-centrin1/CENPA 2:1 cells at anaphase onset treated with or without the Aurora-B inhibitor ZM1. Data from Figure 1E without Aurora-B inhibition are shown for comparison. Aurora-B inhibition allows cells to enter anaphase with asymmetric spindles (n = 12 cells; Mann–Whitney U test, p = 7.57 × 10−6). (C) Distribution of spindle ratios R in Sas-6-depleted 2:1, KIF2a/MCAK-depleted, and KIF2a/MCAK/Sas-6-depleted 2:1 eGFP-centrin1/CENPA cells in metaphase or at anaphase onset. (D) Boxplots of anaphase timing of wild-type, KIF2a/MCAK-depleted, or KIF2a/MCAK/Sas-6 depleted 2:2 and 2:1 eGFP1-centrin2/H2B-mRFP cells. n = 23–61 cells in 1–3 experiments. (E) Quantification of SKAP-negative kinetochores as in Figure 4F in Sas-6-depleted 2:1 and KIF2a/MCAK/Sas-6-depleted 2:1 eGFP-centrin1/CENPA cells. SAC, spindle assembly checkpoint.
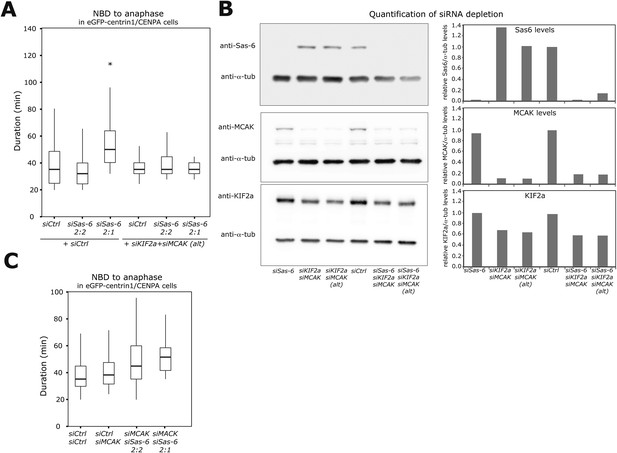
Validation of Sas-6, KIF2a, and MCAK co-depletion.
(A) Boxplots of anaphase timing of siControl, siSas-6 2:2, siSas-6 2:1, siCtrl/KIF2a/MCAK, siKIF2a/MCAK/Sas-6 2:2, and siKIF2a/MCAK/Sas-6 2:1 eGFP1-centrin1/CENPA cells. n = 14–78 cells in 3 independent experiments. Note that siSas-6 2:1 cells are delayed compared to siSas-6 2:2 cells (p < 0.00001 in Mann–Whitney test), but that siKIF2a/MCAK/Sas-6 2:1 are not delayed compared to siKIF2a/MCAK/Sas-6 2:2 cells. (B) Immunoblots of eGFP1-centrin2/H2B-mRFP cells treated with the indicated siRNA and probed with anti-Sas6, anti-MCAK, anti-KIF2A and anti-α-tubulin (loading control) antibodies. The relative ratio with the α-tubulin signal is quantified for each condition on the right panels. Note the KIF2a siRNA only led to a partial depletion. (C) Boxplots of anaphase timing of siControl, siCtrl/MCAK, siMCAK/Sas-6 2:2, siMCAK/Sas-6 2:1 eGFP1-centrin1/CENPA cells. n = 9–169 cells in 3 independent experiments.
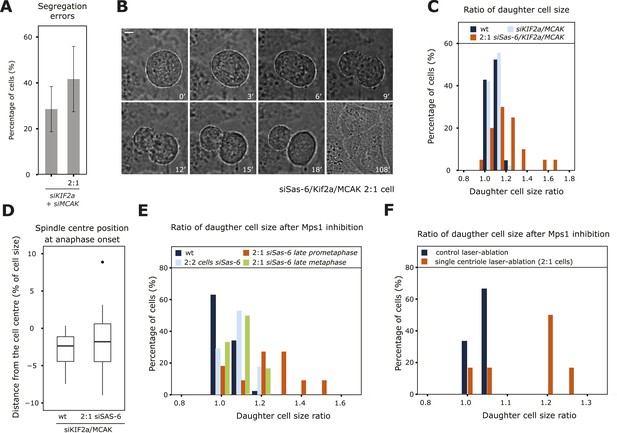
An asymmetric plate position at anaphase onset leads to segregation errors and asymmetric cell division.
(A) Quantification of chromosome segregation errors in KIF2a/MCAK-depleted and KIF2a/MCAK/Sas-6-depleted 2:1 eGFP-centrin1/eGFP-CENPA cells based on time-lapse images. n = 12–21 cells in 2–4 experiments. Error bars indicate s.e.m. (B, C) Wild-type, KIF2a/MCAK-depleted or KIF2a/MCAK/Sas-6-depleted 2:1 eGFP-centrin1/CENPA cells were recorded by time-lapse imaging using the eGFP-centrin1 signal to count centrioles and phase contrast to detect the cell membrane as shown in B for a siKIF2a/MCAK/Sas-6 2:1 cell (scale bar = 5 μm). Phase contrast images were used to quantify the ratio of the two daughter cell sizes, which was plotted as a histogram in C. Half the Kif2a/MCAK/Sas-6-depleted 2:1 cells had a ratio of over 1.2, a ratio never observed in other conditions (t-test with Welch's correction between KIF2a/MCAK and KIF2a/MCAK/Sas-6 2:1, p = 3.1e-05; n = 20–45 cells in 2–4 experiments). (D) Quantification of spindle center position in relation to cell center in KIF2a/MCAK-depleted or KIF2a/MCAK/Sas-6 2:1 eGFP-centrin1/CENPA cells at anaphase onset. n = 20–45 cells in 2–4 experiments. (E) Wild-type HeLa eGFP-centrin1/eGFP-CENPA cells, 2:2 cells, 2:1 cells in late prometaphase (still 1–2 chromosomes not perfectly aligned on the plate), or 2:1 cells in late metaphase (plate perfectly in the middle) were treated with an Mps1 inhibitor and recorded by time-lapse imaging using phase contrast to detect the cell membrane as shown in B. Shown is the ratio of the two daughter cell sizes; 45% of the 2:1 cells treated in late prometaphase/early metaphase had a ratio of over 1.2, a ratio never observed in other conditions (t-test with Welch's correction between 2:1 cells in late prometaphase and 2:1 cells in late metaphase, p = 0.0141; n = 11–17 cells in 3 experiments). See also Videos 5, 6. (F) HeLa eGFP-centrin1/CENPA cells were treated with a laser pulse in the cytoplasm (control) or ablation of a centriole (2:1 cells) and acutely treated with an Mps1 inhibitor to force cells into anaphase. Shown is the ratio of the two daughter cell sizes. Note that 67% of the 2:1 cells treated in late prometaphase/early metaphase had a ratio of over 1.2, a ratio never observed in other conditions (t-test with Welch's correction between control and 2:1 cells p = 0.0451; n = 6). See also Videos 7, 8.
Videos
Sas-6-depleted 2:2 HeLa cell expressing eGFP-centrin1 (centriole marker; green) and mRFP-α-tubulin (microtubules; red) in mitosis.
Time is indicated in minutes.
Sas-6-depleted 2:1 HeLa cell expressing eGFP-centrin1 (centriole marker; green) and mRFP-α-tubulin (microtubules; red) in mitosis.
Time is indicated in minutes. Note the spindle rotation movements.
Sas-6-depleted 1:1 HeLa cell expressing eGFP-centrin1 (centriole marker; green) and mRFP-α-tubulin (microtubules; red) in mitosis.
Time is indicated in minutes.
Laser-ablated 2:1 HeLa cell expressing eGFP-centrin1 (centriole marker) and eGFP-CENPA (kinetochore marker) in metaphase.
Note the asymmetric metaphase plate position after the ablation of a single centriole.
KIF2a/MCAK-depleted HeLa cell expressing eGFP-centrin1 (centriole marker; green) and eGFP-CENPA (kinetochore marker; green) entering anaphase and recorded with phase contrast microscopy.
https://doi.org/10.7554/eLife.05124.017KIF2a/MCAK/Sas-6-depleted 2:1 HeLa cell expressing eGFP-centrin1 (centriole marker; green) and eGFP-CENPA (kinetochore marker; green) entering anaphase and recorded with phase contrast microscopy.
Note the asymmetric cell division.
Laser-ablated control (ablation in the cytoplasm) HeLa cell expressing eGFP-centrin1 (centriole marker) and eGFP-CENPA (kinetochore marker) treated in metaphase with an Mps1 inhibitor.
Shown is the GFP-fluorescence channel (left) and the DIC channel (right). Note how the cell divides in a symmetric manner.
Laser-ablated 2:1 HeLa cell expressing eGFP-centrin1 (centriole marker) and eGFP-CENPA (kinetochore marker) treated in metaphase with an Mps1 inhibitor.
Shown is the GFP-fluorescence channel (left) and the DIC channel (right). Note how the cell divides in an asymmetric manner.
Tables
Number of cells in every experiment
| Condition | N° of cells |
|---|---|
| WT | 40 |
| WT at anaphase onset | 42 |
| WT + MPS1-IN | 19 |
| SiSas-6 2:2 cells | 41 |
| SiSas-6 2:2 cells at anaphase onset | 29 |
| SiSas-6 2:2 cells + MPS1-IN | 19 |
| SiSas-6 2:1 cells | 59 |
| siSas-6 2:1 cells + MG132 | 18 |
| SiSas-6 2:1 cells at anaphase onset | 33 |
| SiSas-6 2:1 cells in long term videos | 14 |
| SiSas-6 2:1 cells + MPS1-IN | 26 |
| SiSas-6 1:1 cells | 36 |
| SiKif2a + siMCAK | 20 |
| SiKif2a + siMCAK at anaphase onset | 41 |
| SiKif2a + siMCAK + siSas-6 2:1 cells | 29 |
| SiKif2a + siMCAK + siSas-6 2:1 cells at anaphase onset | 24 |
| SiSas-6 2:1 cells + ZM1 | 10 |
| SiSas-6 2:2 + DMSO | 15 |
| SiSas-6 2:2 + taxol | 11 |
| SiSas-6 2:1 + DMSO | 16 |
| SiSas-6 2:1 + taxol | 15 |
| Centriole laser-ablation (2:1) | 11 |
| Control laser-ablation | 8 |
| Centriole laser-ablation (2:1) + Mps1 inhibitor | 6 |
| Control laser-ablation + Mps1 inhibitor | 6 |
Additional files
-
Source code 1
Custom built software in Matlab.
- https://doi.org/10.7554/eLife.05124.021





