The zinc finger proteins ZNF644 and WIZ regulate the G9a/GLP complex for gene repression
Figures
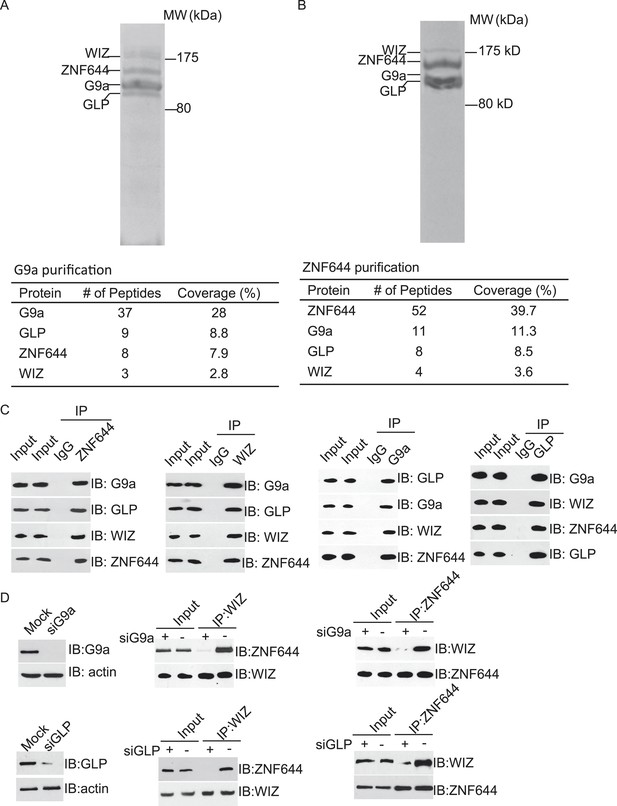
ZNF644 and WIZ associate with G9a.
(A) Silver staining of affinity-purified G9a complex. Cell lysates of 293T cells stably expressing SFB-G9a were subjected to affinity purification. Eluted proteins were visualized by silver staining. Arrows indicate proteins corresponding to G9a, GLP, ZNF644 and WIZ. Peptide coverage is shown in the table. (B) Silver staining of affinity-purified ZNF644 partners. (C) ZNF644 and WIZ co-exist in the same complex with G9a and GLP. U2OS cell lysates were analyzed by co-immunoprecipitation (co-IP) and Western blotting with the antibodies indicated. The whole cell lysates of U2OS was used as the input. An irrelevant IgG was used as the IP control. (D) Down-regulation of G9a or GLP impairs the interaction between WIZ and ZNF644. G9a and GLP were down-regulated by siRNAs in U2OS cells. The cell lysates were analyzed by IP and Western blotting with the antibodies indicated.
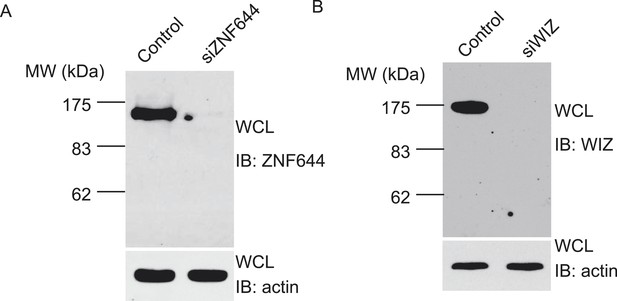
ZNF644 and WIZ antibodies have been generated and specifically recognize the endogenous ZNF644 and WIZ respectively.
(A) Anti-ZNF644 antibody recognizes endogenous ZNF644 from U2OS cell lysates. In cell lysates, the antibody specifically recognized a band around 150 kD. When cells were treated with siZNF644, this band was disappeared. Anti-actin was used as protein loading control. (B) Anti-WIZ antibody recognizes endogenous WIZ from U2OS cell lysates. In cell lysates, the antibody specifically recognized a band around 175 kD.
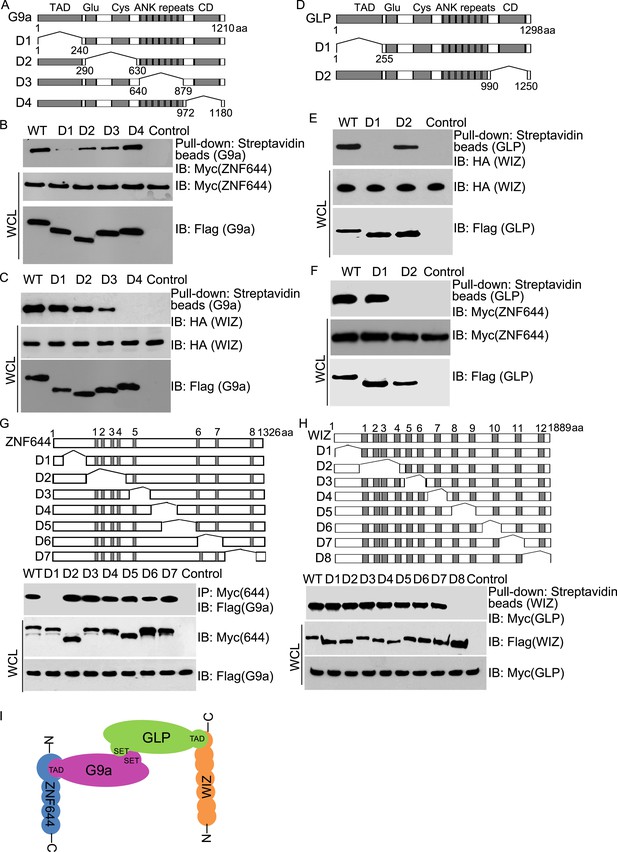
Mapping the interaction regions of ZNF644, WIZ, G9a and GLP.
(A) A series of deletion mutants of SFB-tagged G9a were generated to map the interaction region of G9a. CD: catalytic domain. (B) The D1 mutant of G9a abolishes the interaction with ZNF644. SFB-tagged wild-type G9a and deletion mutants were expressed in 293T cells together with Myc-ZNF644. The cell lysates were subjected to streptavidin beads pull-down and Western blotting with the indicated antibodies. The whole cell lysates were used as the input. Cells only expressing Myc-ZNF644 were used for pull-down control. (C) Lacking the catalytic domain of G9a (CD) disrupts the interaction with WIZ. (D) The TAD and catalytic domain deletion mutants of GLP are generated. (E) The TAD domain of GLP is important for the interaction with WIZ. (F) Lacking the catalytic domain of GLP abolishes the interaction with ZNF644. (G) The N-terminus deletion mutant of ZNF644 abolishes the interaction with G9a. Myc-tagged ZNF644 and deletion mutants were co-expressed together with SFB-G9a in 293T cells. IP and Western blotting were performed with indicated antibodies. (H) The C-terminus deletion mutant of WIZ abolishes the interaction of WIZ and GLP. SFB-tagged WIZ and deletion mutants were co-expressed with Myc-GLP in 293T cells. (I) A model shows that the N-terminus of ZNF644 interacts with the TAD of G9a, while the C-terminus of WIZ interacts with the TAD of GLP.

Down-regulation of G9a doesn’t affect the interaction between WIZ and GLP.
(A) The siRNA targeting G9a is used to down-regulate G9a in U2OS cells. (B) The cell lysates were analyzed by co-IP and Western blotting with indicated antibodies.
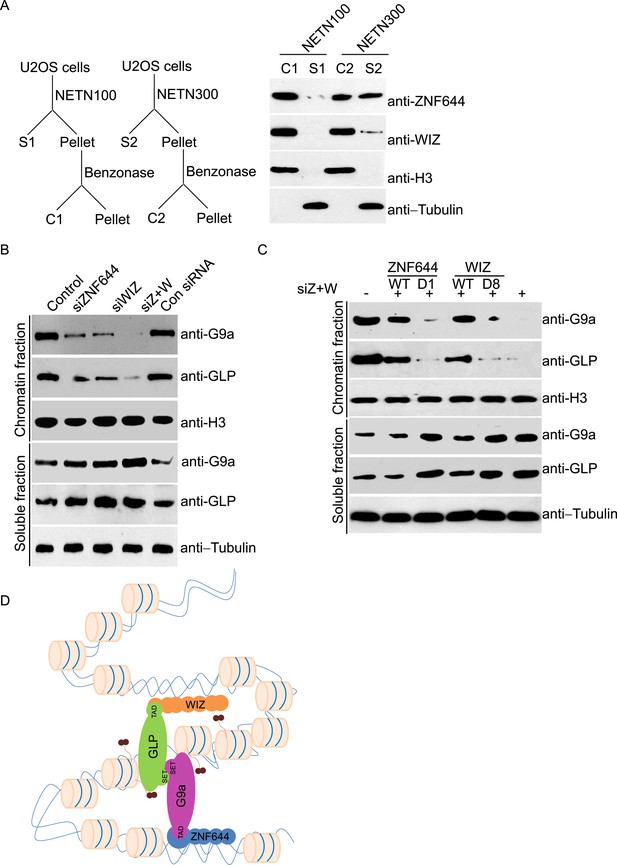
ZNF644 and WIZ are important for the chromatin localization of G9a.
(A) Both ZNF644 and WIZ tightly bind to chromatin. U2OS cells were lysed by NETN100 (lysis buffer with 100 mM NaCl) and NETN300 (lysis buffer with 300 mM NaCl) respectively. After harvesting the soluble fractions, the pellets were digested by Benzonase to extract the chromatin fraction. Each fraction was examined by Western blotting. Tubulin and histone H3 were used as loading control for the soluble fraction and chromatin fraction respectively. (B) Knockdown of ZNF644 or/WIZ impairs the chromatin association of G9a and GLP. U2OS cells were lysed with NETN100 buffer. The soluble fraction and chromatin fraction were separated and each fraction was examined with Western blotting. Tubulin and histone H3 were used as loading control in the soluble fraction and chromatin fraction respectively. (C) In the cells with siRNA-resistant ZNF644 or WIZ, G9a is retained in the chromatin fraction. But the D1 mutant of ZNF644 or the D8 mutant of WIZ is unable to target G9a to chromatin. (D) A model shows that ZNF644 and WIZ facilitate the chromatin localization of the G9a/GLP complex.
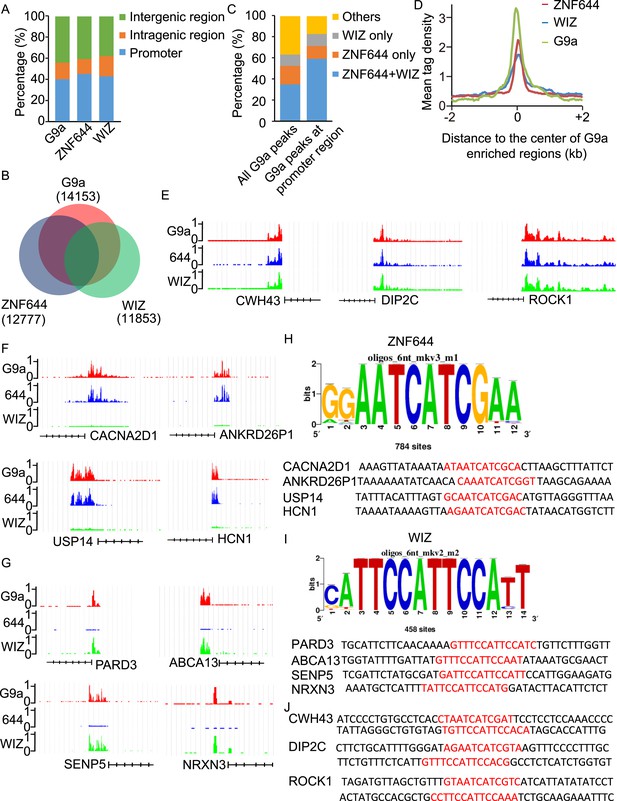
WIZ and ZNF644 associate with G9a at specific genomic loci.
(A) Summary of genome-wide distribution of G9a, ZNF644 and WIZ in different regions. Y-axes: percentage of each region in the genome. (B) Venn diagram shows a significant overlap between G9a, ZNF644 and WIZ enriched peaks. (C) The G9a-enriched peaks were bound with ZNF644 and/or WIZ, especially in promoter region. (D) G9a, ZNF644 and WIZ ChIP-seq read counts in 100-bp window were plotted against the distance (−2 kb, +2 kb) from the center of G9a enriched regions in promoter region. Y-axes: mean tag density. (E) ChIP-seq results show the co-occupancy of ZNF644, WIZ and G9a at CWH43, DIP2C and ROCK1 loci. (F) ZNF644 and G9a are co-localized at the promoter regions of CACNA2D1, ANKRD26P1, USP14 and HCN1. (G) WIZ and G9a are co-localized at the promoter regions of PARD3, ABCA13, SENP5 and NRXN3. (H) The consensus DNA-binding motif of ZNF644 is analyzed according to ChIP-seq result. The binding sequences in CACNA2D1, ANKRD26P1, USP14 and HCN1 loci are shown in red. (I) The specific DNA binding sequence of WIZ is obtained according to the ChIP-seq results, and is confirmed at PARD3, ABCA13, SENP5 and NRXN3 loci. (J) Both ZNF644 and WIZ-binding sequences are identified at CWH43, DIP2C and ROCK1 loci, which are co-occupied by ZNF644 and WIZ.
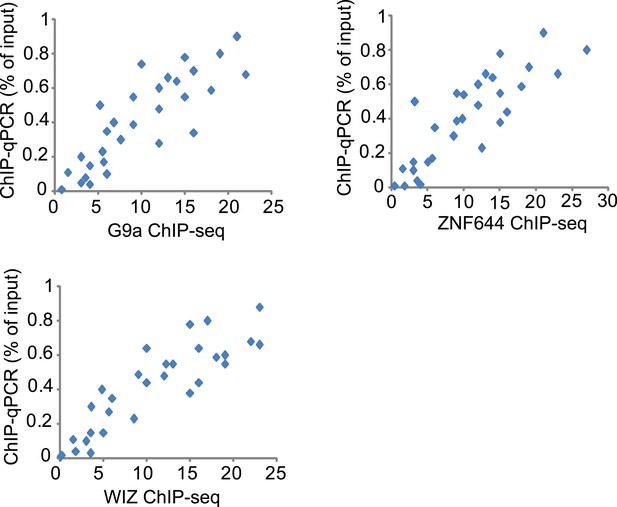
Validation of ChIP-seq results by qPCR.
ChIP-seq fragment densities of G9a (x-axis) are plotted against ChIP-qPCR fold-enrichment of G9a (percentage of input) (y-axis) at 30 selected loci in 293T cells that represent a broad range of ChIP-seq fragment counts. The 30 selected loci contain 10 loci that are G9a positive (ChIP-seq signal >5), 10 loci that are G9a, ZNF644 and WIZ positive, and the other 10 loci are G9a negative. The same methods were used to analyze the ChIP-seq result of ZNF644 and WIZ. 20 loci identified as significantly enriched by ChIP-seq were clearly different from 10 unenriched loci in the plots.
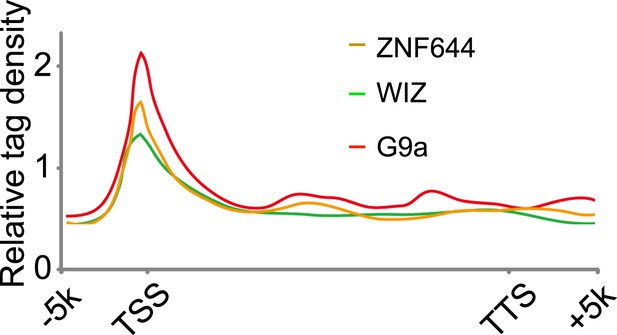
Genome-wide analysis of ChIP-seq peaks.
Average genome-wide occupancies of G9a, ZNF644 and WIZ along the transcription unit. TSS and TES, the transcription start and end sites, respectively.
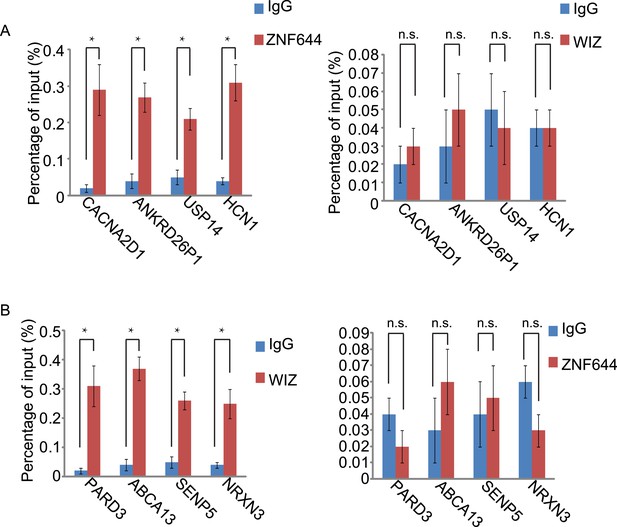
The gene loci occupied by ZNF644 or WIZ are confirmed by ChIP-qPCR.
(A) ChIP-qPCR confirms the occupancy of ZNF644, but not WIZ, at CACNA2D1, ANKRD26P1, USP14 and HCN1loci. *p < 0.05; n.s., not significant. (B) The occupancy of WIZ, but not ZNF644, is shown at PARD3, ABCA13, SENP5 and NRXN3 loci. *p < 0.05; n.s., not significant.
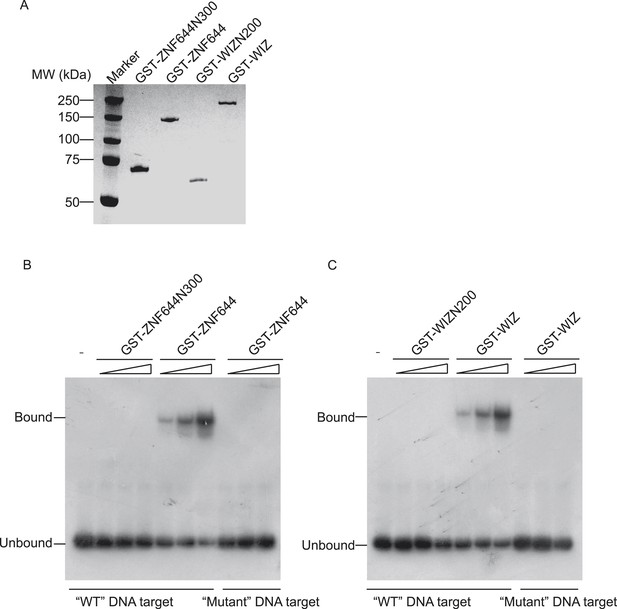
DNA binding motifs of ZNF644 or WIZ concluded from ChIP-seq results were validated by Electrophoretic Mobility Shift Assay (EMSA).
(A) GST-tagged full length ZNF644 (GST-ZNF644), N-terminus of ZNF644 (a.a. 1-300) (ZNF644N300), full length human WIZ (GST-WIZ) or N-terminus of WIZ (a.a. 1-200) (WIZN200) were purified from Sf9 insect cells and used for EMSA. The proteins were purified by GST beads and examined by coomassie blue staining. (B) Recombinant GST-ZNF644 or N terminus of ZNF644 without Zinc finger motif (GST-ZNFN300) was incubated with 32P-labeled 48-mer sequence motif-contained DNA oligonucleotides. Only GST-ZNF644, but not ZNF644N300, could bind to the DNA containing sequence motif. The 32P-labeled 48-mer DNA oligonucleotides containing “mutant” DNA target was used as the negative control. (C) Recombinant GST-WIZ or N terminus of WIZ without Zinc finger motif (GST-WIZN200) was incubated with 32P-labelled 48-mer sequence motif-contained DNA oligonucleotides. Only GST-WIZ, but not GST-WIZN200, could bind to the DNA containing consensus motif. The 32P-labeled 48-mer DNA oligonucleotides containing “mutant” DNA target was used as the negative control.
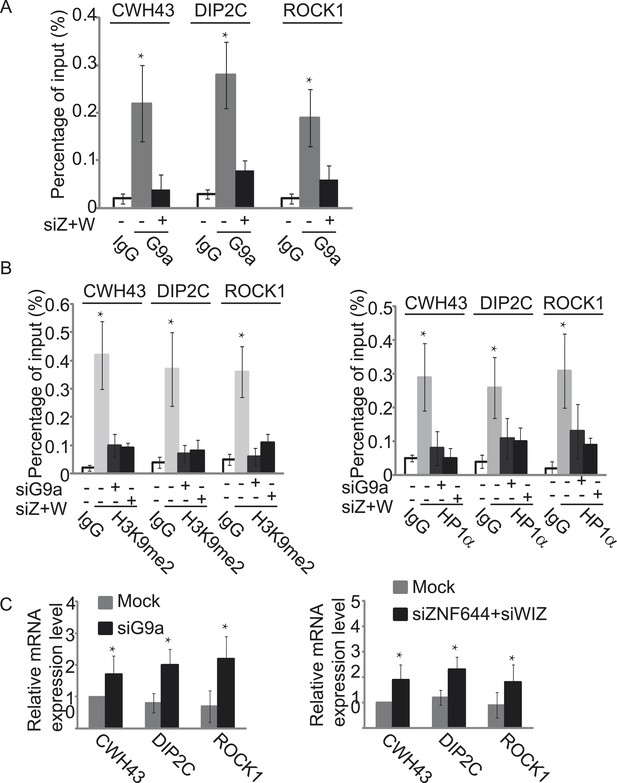
ZNF644 and WIZ target G9a for gene repression.
(A) Down-regulation of ZNF644 and WIZ by siRNAs (siZ + W) impairs the localization of G9a at CWH43, DIP2C and ROCK1 loci. *p < 0.05 compared to IgG. (B) Knockdown G9a abolishes the enrichment of H3K9me2 and HP1α at CWH43, DIP2C and ROCK1 loci. Loss of ZNF644 and WIZ also impairs the enrichment of H3K9me2 and HP1α at these loci. *p < 0.05 compared to IgG. (C) Down-regulation of G9a by siRNA increases gene transcription at CWH43, DIP2C and ROCK1 loci, and loss of ZNF644 and WIZ also facilitates gene transcription at these loci. *p < 0.05 compared to Mock.
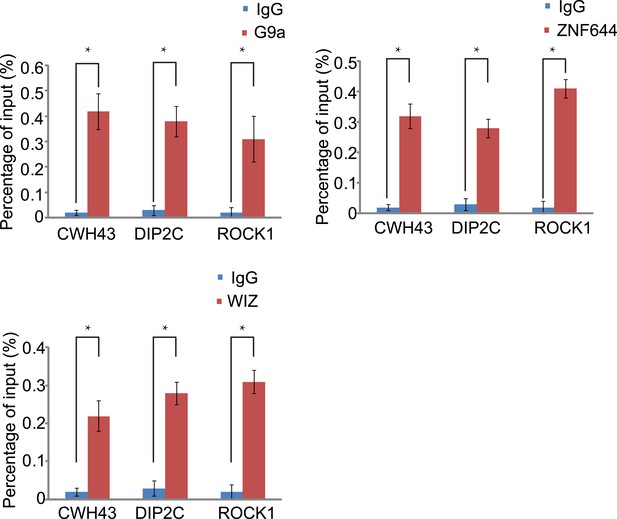
Co-occupancy of ZNF644, WIZ and G9a is shown at CWH43, DIP2C and ROCK1 loci.
ChIP-qPCR was performed to confirm the ChIP-seq results. *p < 0.05.
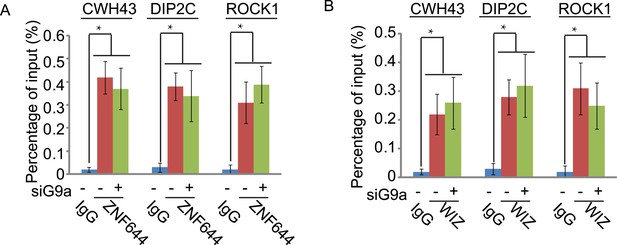
Down-regulation of G9a does not affect the chromatin localization of ZNF644 (A) and WIZ (B) at CWH43, DIP2C and ROCK1 loci.
ChIP-qPCR was performed in the siG9a-treated U2OS cells. *p < 0.05.
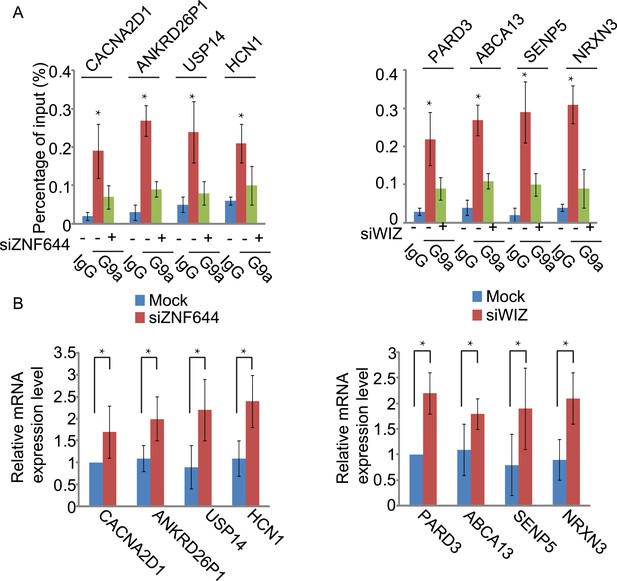
ZNF644 and WIZ target G9a for gene repression at only ZNF644 or WIZ occupied gene loci.
(A) Knockdown ZNF644 by siZNF644 impairs the recruitment of G9a at CACNA2D1, ANKRD26P1, USP14 and HCN1 loci. Knockdown WIZ abolishes the localization of G9a at PARD3, ABCA13, SENP5 and NRXN3 loci. *p < 0.05 compared to IgG. (B) Knockdown ZNF644 facilitates the gene transcription at CACNA2D1, ANKRD26P1, USP14 and HCN1 loci, while knockdown WIZ induces the gene transcription at PARD3, ABCA13, SENP5 and NRXN3 loci. *p < 0.05 compared to Mock.
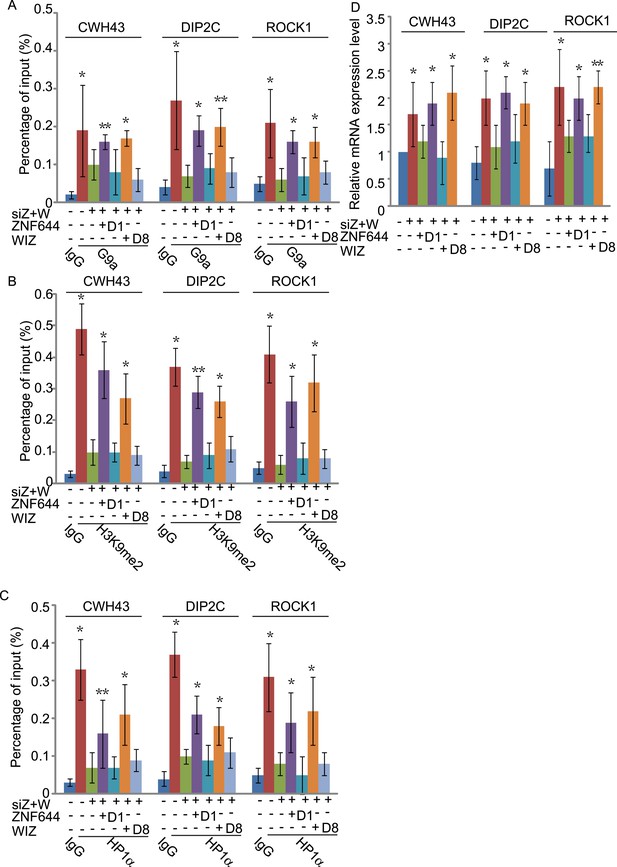
The interaction domains in ZNF644 and WIZ are important for the G9a-dependent function.
(A) Wild-type ZNF644 and WIZ, but not the D1 mutant of ZNF644 and the D8 mutant of WIZ, rescue the recruitment of G9a to CWH43, DIP2C and ROCK1 loci. *p < 0.05, **p < 0.01 compared to IgG. (B-D) Wild-type ZNF644 and WIZ, but not the D1 mutant of ZNF644 and the D8 mutant of WIZ, restore the enrichment of H3K9me2 and HP1α as well as gene transcription at CWH43, DIP2C and ROCK1 loci. *p < 0.05, **p < 0.01 compared to IgG (B, C) or the control U2OS cells without siRNAs treatment (D).
Additional files
-
Supplementary file 1
PCR primers. All the primers used for RT-PCR and ChIP-qPCR are listed.
- https://doi.org/10.7554/eLife.05606.018





