Cdc6 ATPase activity disengages Cdc6 from the pre-replicative complex to promote DNA replication
Figures
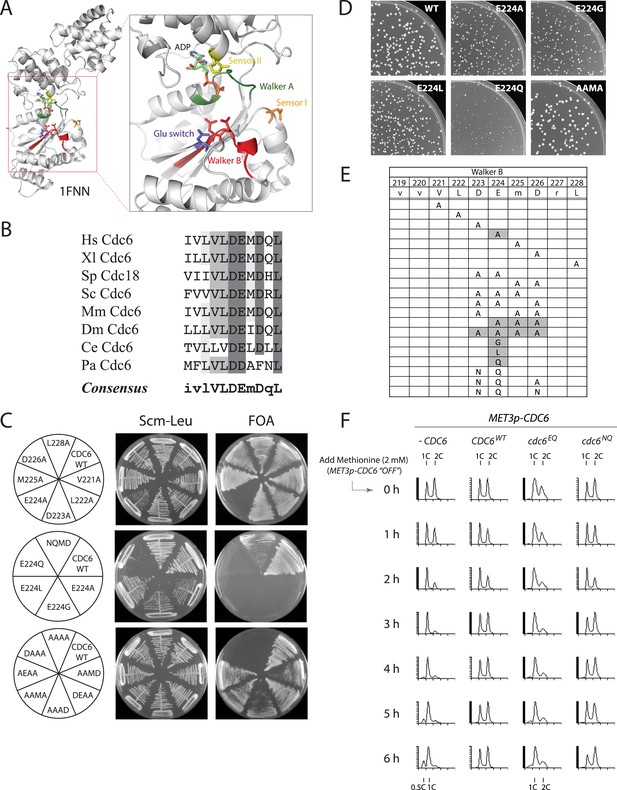
The Cdc6 Walker B catalytic residue is essential in yeast but is easily bypassed by intragenic suppressor mutations.
(A) Structure of the Pyrobaculum aerophilum Cdc6 orthologue with bound ADP (Liu et al., 2000) highlighting the ATP binding pocket and key residues. The ‘DD’ Walker B residues are colored red and the sensor I residue is colored orange. (B) Multiple sequence alignment of Cdc6 Walker B motif. Uppercase letters in consensus are highly conserved. From top to bottom: Homo sapiens, Xenopus laevis, S. pombe, S. cerevisiae, Mus musculus, Drosophila melanogaster, C. elegans, and Pyrobaculum aerophilum Cdc6 homologues. (C) Viability analysis of Cdc6 Walker B motif mutants reveals only E224 is essential. M4466 (cdc6Δ::ura3 pRS416-CDC6) was transformed with the indicated pMW71-derived plasmids (listed in Supplementary file 2) and then restreaked onto SCM-Leu plates representing growth of CDC6/cdc6 or FOA plates, growth of cdc6 mutant only. (D) Growth of plasmid cdc6 derivatives transformed into wild-type yeast (W303-1A) reveals that multiple E224 substitutions have dominant growth affects. (E) Summary of Cdc6 mutational analysis. Shaded regions are non-viable mutants. (F) Flow cytometry profiles of strains M4759 (K4055 × pRS415, vector), M4758 (K4055 × pMW71, CDC6-WT), M4760 (K4055 × pFJ21, cdc6-EQ), and M4762 (K4055 × pFJ230, cdc6-NQ) after addition of methionine to repress expression of wild type MET3p-CDC6 present in all strains.
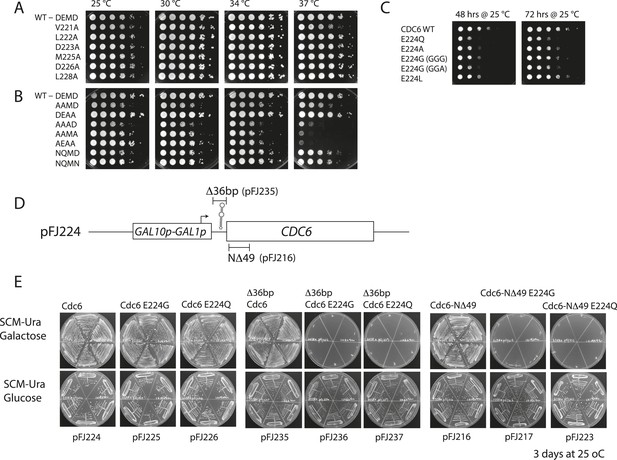
Growth properties of various cdc6 Walker B mutants.
For (A) and (B), M4466 (W303-1A, cdc6Δ::ura3) containing only the indicated cdc6 alleles on pMW71 (Supplementary file 2) were spotted onto YPD plates in a 10-fold dilution series and incubated at the indicated temperatures. (A) Alanine scanning mutants across the Cdc6 Walker B box. (B) Double and triple mutants within the ‘DEMD’ core region. The AAAD, AAMA, and AEAA mutants exhibited substantial temperature sensitivity. (C) The indicated Cdc6-E224 mutant alleles or wild-type CDC6 on pMW71 (Supplementary file 2) were transformed into wild-type yeast (W303-1A). These transformants were spotted using 10-fold serial dilutions onto SCM-Leu plates (selecting for the pMW71-derivative) to quantitatively measure their dominant negative growth phenotype. This indicates that all the E224 alleles exhibited some dominate growth effects over the wild type. Two separate codons for E224G were tested. (D) Diagram of pGAL-CDC6 plasmids. pFJ224 contains the GAL1,10 promoter 39 bp upstream of wild-type CDC6 coding sequence to give a GAL1p-CDC6 promoter fusion. This plasmid contains a 49-bp sequence (CCGGGAATTTCCGGTGGTGGTGGTGGAATTCTAGACTCC ATG TCA GCT A) predicted to form a stable multi-stem loop structure that overlaps the Cdc6 ATG, underlined. The 39 bp immediately preceding the Cdc6 ATG is derived from vector sequences. This pGAL-Cdc6 construct complements Cdc6 function when yeast is propagated on galactose but not on glucose media. (E) Overexpression of Cdc6-E224G or -E224Q mutant proteins from pFJ216 (differing from pFJ224 only by a deletion of Cdc6 residues 2–49, which significantly stabilizes Cdc6 protein) or from pFJ235 (differing from pJF224 only by a 36 bp deletion disrupting the stem loop) on galactose causes complete growth inhibition of wild type yeast. In contrast, galactose-induced expression of these mutant proteins in the pFJ224 plasmid background causes only a mild growth inhibition, indicative of lower Cdc6 protein induction. The indicated plasmids (Supplementary file 2) were transformed into wild-type yeast (W303-1A), and 6 transformants each were streaked onto selective minimal media (SCM-Ura) containing galactose or glucose as the carbon source and incubated at 25°C for 3 days.
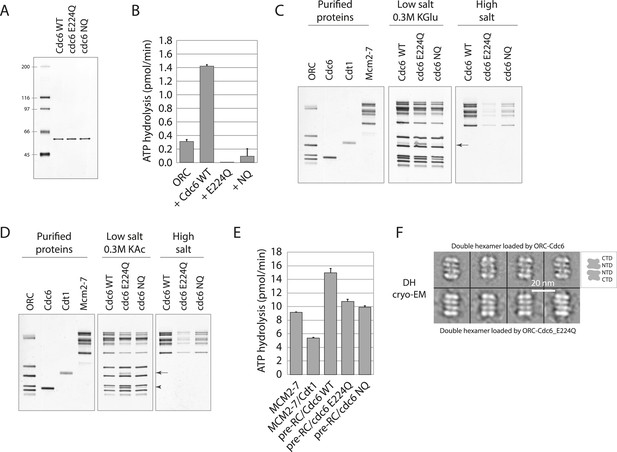
Cdc6 ATPase activity is not required for MCM loading in vitro.
(A) Silver-stained 10% SDS gel with molecular weight standards and 100 ng of the indicated purified Cdc6 proteins. (B) Cdc6 ATPase assays. (C) MCM loading assay using the purified proteins shown on left. ‘Low salt’ shows proteins associated with DNA, ‘High salt’ wash reveals loaded MCM protein. Arrow marks Cdt1. (D) MCM loading assay as in (C) using a more stringent (0.3 M K acetate) low salt wash indicates that Cdc6-E224Q is stabilized on DNA relative to WT Cdc6 and Cdc6-NQ protein. Arrow marks Cdt1, arrowhead marks Cdc6. (E) ATPase assays with Cdc6-WT-ORC-DNA, Cdc6-E224Q-ORC-DNA, or Cdc6-NQ-ORC-DNA complexes after MCM-Cdt1 addition. The first two lanes show soluble MCM and MCM-Cdt1 ATPase activities as controls. (F) The double hexamer of Mcm2-7 loaded by Cdc6-E224Q after high salt wash (1 M NaCl) of loading reactions is indistinguishable from that loaded by wild-type Cdc6. 528 raw cryo-EM particle images were used for 2D classification and averaging for the dhMCM protein loaded with ORC-Cdc6-E224Q. 3217 raw particles were used to generate the averaged views of the dhMCM loaded with wild-type ORC-Cdc6.
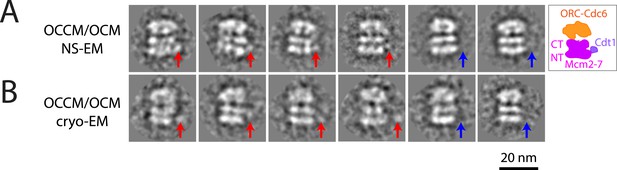
MCM loading by ORC-Cdc6-E224Q gives rise to a heterogeneous mixture of intermediates by EM.
(A) Negative stain-EM and (B) cryo-EM of complexes formed with ORC, Cdc6-E224Q, and MCM-Cdt1 proteins revealed a heterogeneous mixture of OCCM (ORC-Cdc6-Cdt1-MCM) and OCM (ORC-Cdc6-MCM) complexes; averages from 3655 individual particle images and 1436 cryo-EM raw particle images, respectively. The red arrows point to Cdt1 density, and blue arrows indicate the absence of Cdt1. The structure of Cdc6-E224Q-containing OCCM particles is similar to wild-type OCCM assembled in the presence of ATP-γ-S. However, the Cdt1 density is variable in its location in the mutant OCCM and is of course, absent in OCM complexes. Most of the particles were OCCM. We do not know the exact ratio between OCCM and OCM because the views without Cdt1 density are not necessarily of OCM; they could be OCCM particles at slightly different side views that prevented visibility of Cdt1.
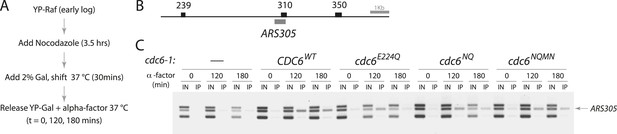
Cdc6 ATPase mutants promote MCM origin binding in yeast.
(A) Cell synchronization protocol for MCM ChIP. (B) PCR products amplified in base pairs surrounding ARS305. (C) All the cdc6 ATPase mutants tested promote MCM binding to origins similar to the wild-type CDC6 regardless of whether they complement yeast viability. Strains used: M378 (cdc6-1), M4455 (cdc6-1 GAL1p-CDC6::LEU2), M4531 (cdc6-1 GAL1p-cdc6-E224Q::LEU2), M4513 (cdc6-1 GAL1p-cdc6-NQ::LEU2) and M4464 (cdc6-1 GAL1p-cdc6-NQMN::LEU2).
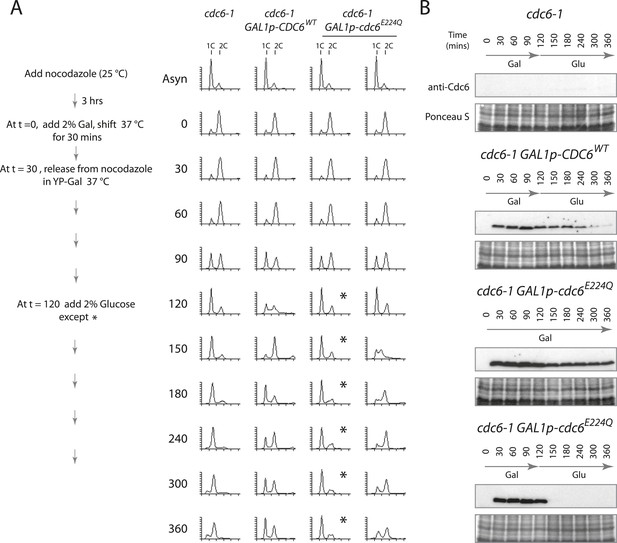
Cdc6 removal after MCM loading bypasses ATPase requirement and is sufficient to allow DNA replication.
(A) Synchronization protocol (left) and flow cytometry profiles (right) of yeast strains M378 (cdc6-1), M4455 (cdc6-1 GAL1p-CDC6::LEU2), and M4531 (cdc6-1 GAL1p-cdc6-E224Q::LEU2). Asynchronous cells were arrested in G2/M for 3 hr with nocodazole, shifted up to 37°C for 30 min (from t = 0 to 30 min), and then released into G1-phase at 37°C expressing no additional Cdc6, GAL1p-CDC6 or GAL1p-cdc6-E224Q. GAL1 promoter-driven CDC6 expression was shut off at t = 120 min by the addition of glucose except where marked by an asterisk. Identical flow profiles to M4531 were seen using an independent cdc6-1 GAL1p-cdc6-E224Q integrant strain, M4530. (B) Cdc6 Western blots (top panels) and total protein (bottom panels) by Ponceau S staining of the samples are shown in panel A. The addition of glucose at 120 min causes Cdc6 wild-type protein to disappear and this occurs more rapidly for the Cdc6-E224Q mutant.
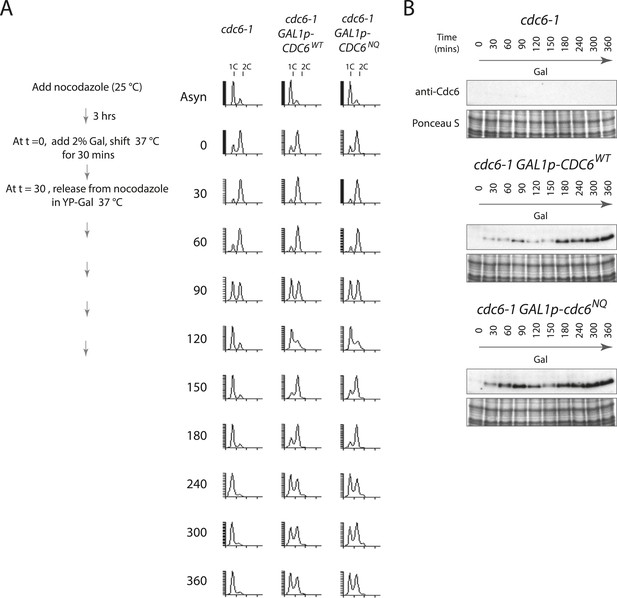
Overproduction of wild-type or Cdc6-NQ proteins does not cause a G1-arrest.
(A) Cell synchronization protocol (right) and flow cytometry profiles (left) of yeast strains M378 (cdc6-1), M4455 (cdc6-1 GAL1p-CDC6::LEU2), and M4513 (cdc6-1 GAL1p-cdc6-NQ::LEU2). Asynchronous cells were arrested in G2/M for 3 hr with nocodazole, shifted up to 37°C for 30 min (from t = 0 to 30 min), and then released into G1-phase at 37°C expressing no additional Cdc6, GAL1p-CDC6, or GAL1p-cdc6-NQ. Cells overexpressing wild-type Cdc6 or Cdc6-NQ proteins proceed normally into S-phase. (B) Cdc6 Western blots (top panels) and total protein (bottom panels) by Ponceau S staining of the samples is shown in panel A.
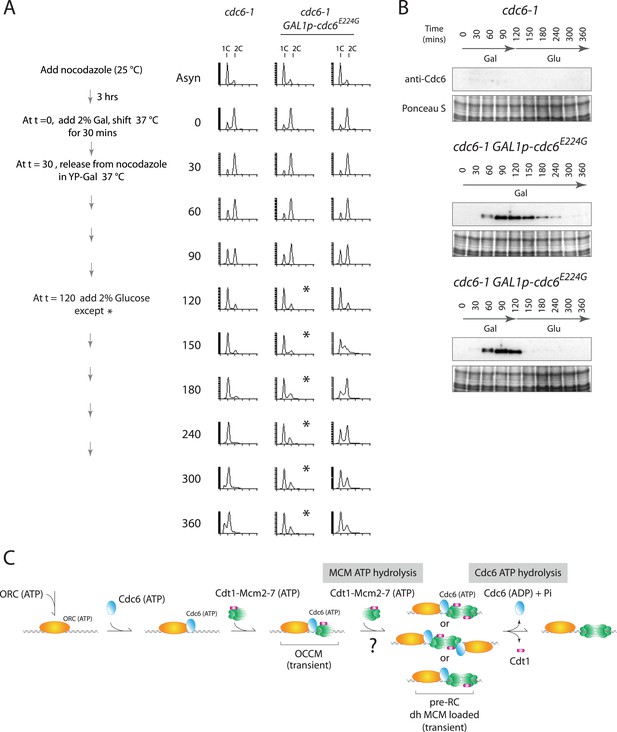
Cdc6-E224G ATPase mutant is also defective in G1 progression but its degradation promotes DNA replication.
(A) Synchronization protocol (left) and flow cytometry profiles (right) of yeast strains M378 (cdc6-1) and M4766 (cdc6-1 GAL1p-cdc6-E224G::LEU2). Asynchronous cells were arrested in G2/M for 3 hr with nocodazole, shifted up to 37°C for 30 min (from t = 0 to 30 min), and then released into G1-phase at 37°C expressing no additional Cdc6 or GAL1p-cdc6-E224G as in Figure 4. (B) Cdc6 Western blots (top panels) and total protein (bottom panels) by Ponceau S staining of the samples shown in panel A. (C) Model for the role of Cdc6 ATP hydrolysis in DNA replication initiation (see text).
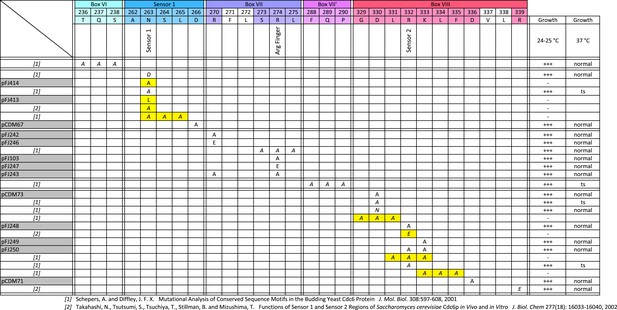
Growth phenotypes of cdc6 mutants altering additional residues that potentially affect ATP hydrolysis or ATP sensing.
Mutational summary of Box VI, sensor 1, Box VII (R-finger), Box VII', and Box VIII (sensor 2) regions of Cdc6. Yellow highlighting indicates that the mutation gave a lethal phenotype in yeast at 25°C. Plasmids from this study or previous published references for each mutant are listed at the left, and growth properties are listed at the right.
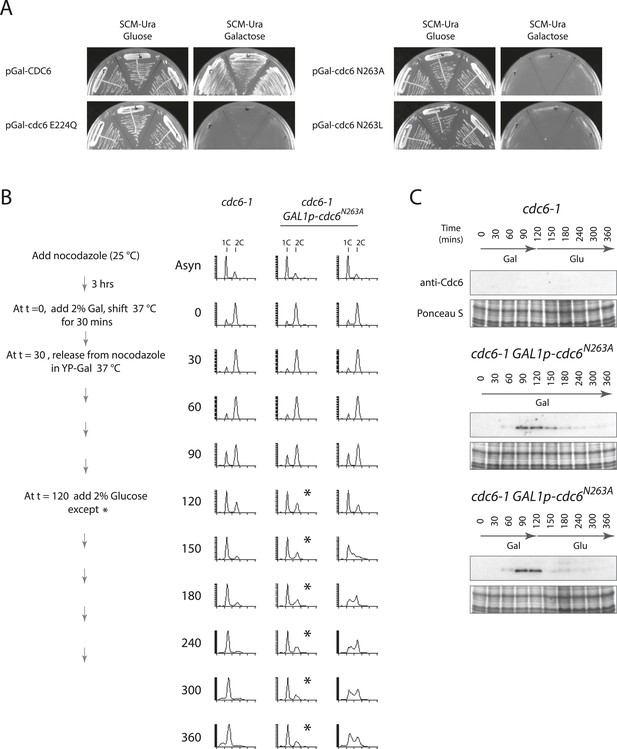
Expression of the Cdc6-N263A ATPase mutant protein is dominant negative for growth, causes a G1 block, but loads functional MCM that can promote DNA replication after Cdc6-N263A degradation.
(A) pFJ235 (pRS416 Gal1p-CDC6) and mutant derivatives pFJ237 (Gal1p-cdc6-E224Q), pFJ404 (Gal1p-cdc6-N263A), and pFJ412 (Gal1p-cdc6-N263L) (Supplementary file 2) were transformed into wild-type yeast, W303-1A. Multiple transformants were streaked onto SCM-Ura plates containing glucose or galactose. (B) Cell synchronization protocol (left) and flow cytometry profiles (right) of yeast strains M378 (cdc6-1) and M4763 (cdc6-1 GAL1p-cdc6-N263A::LEU2). Asynchronous cells were arrested in G2/M for 3 hr with nocodazole, shifted up to 37°C for 30 min (from t = 0 to 30 min), and then released into G1-phase at 37°C expressing no additional Cdc6 or GAL1p-cdc6-N263A. GAL1 promoter-driven cdc6-N263A expression was shut off at t = 120 min by the addition of glucose except where marked by an asterisk. (C) Cdc6 Western blots (top panels) and total protein (bottom panels) by Ponceau S staining of the samples are shown in panel (B).
Additional files
-
Supplementary file 1
Yeast strains used in this study.
- https://doi.org/10.7554/eLife.05795.013
-
Supplementary file 2
Plasmids used in this study.
- https://doi.org/10.7554/eLife.05795.014





