A secretory kinase complex regulates extracellular protein phosphorylation
Figures
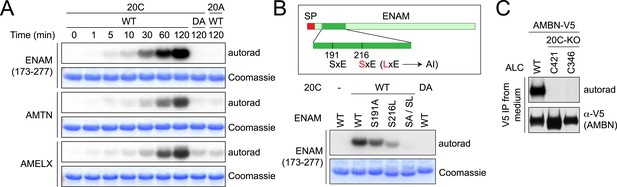
Fam20C phosphorylates the enamel matrix proteins within SxE motifs.
(A) Phosphorylation of enamel matrix proteins by Fam20C in vitro. 250 μg/ml purified recombinant human ENAM (173–277), AMTN or AMELX was incubated with recombinant Fam20C, Fam20C-D478A (DA) or Fam20A (5 μg/ml) at 30°C in the presence of [γ-32P]-ATP. After reaction, proteins were separated by SDS-PAGE and visualized by Coomassie staining. 32P incorporation was detected by autoradiography. (B) Effect of SxE motif mutations on ENAM phosphorylation by Fam20C. Upper: the schematic of ENAM protein. Dark green, ENAM (173–277). SP, signal peptide; AI, Amelogenesis Imperfecta. S216L mutation was reported to cause AI. Lower: phosphorylation of different forms of ENAM (173–277) by Fam20C. SA/SL, S191A and S216L. (C) Effect of Fam20C KO on the phosphorylation of AMBN. C-terminal V5-tagged AMBN (AMBN-V5) was expressed in WT ALCs or two Fam20C-KO ALC clones (C421 and C346) that were metabolically labeled with 32P orthophosphate. AMBN-V5 was immunoprecipitated from the conditioned media. Total protein and 32P incorporation were detected by western blot and autoradiography, respectively.
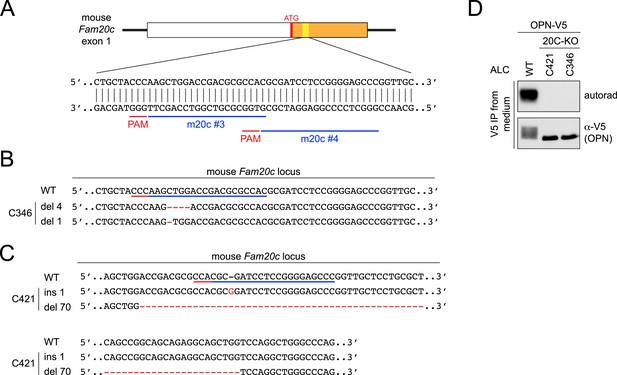
CRISPR-mediated Fam20C KO in ALCs and its effect on osteopontin (OPN) phosphorylation.
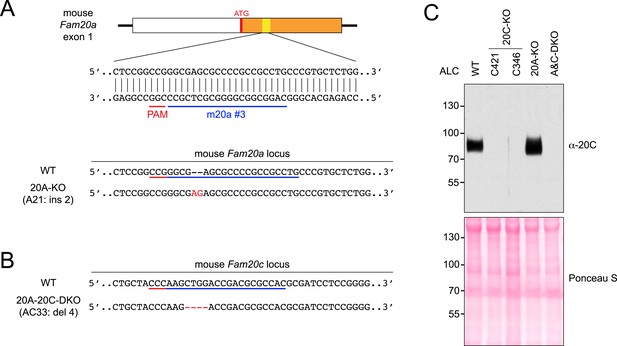
(A) The schematic of the single-guide (sg)RNA-targeting sites in the mouse Fam20c gene. Two targeting sequences were used in this study: m20c #3 and #4. The first exon of the mouse Fam20c gene is shown as a box. White, 5′ UTR; Orange, the coding sequence. Targeting sites and protospacer adjacent motifs (PAMs) are indicated as blue and red bars, respectively. (B and C) Sequence alignment of WT Fam20c gene and the disrupted alleles from Fam20C-KO ALC clone C346 (B) or C421 (C). Targeting sites and PAMs are indicated as blue and red bars, respectively. Inserted nucleotides are highlighted in red. Deleted regions are indicated with red dashes. Del X, deletion of X nucleotides; ins X, insertion of X nucleotides. (D) Effect of Fam20C KO on OPN phosphorylation. OPN is a model substrate of Fam20C. C-terminal V5-tagged OPN (OPN-V5) was expressed in WT ALC or two Fam20C-KO ALC clones (C346 or C421) that were metabolically labeled with 32P orthophosphate. OPN-V5 was immunoprecipitated from the conditioned media. Total protein and 32P incorporation were detected by western blot and autoradiography, respectively. No phosphorylation of OPN-V5 was detected in the KO cells, indicating that Fam20C activity was completely abolished.
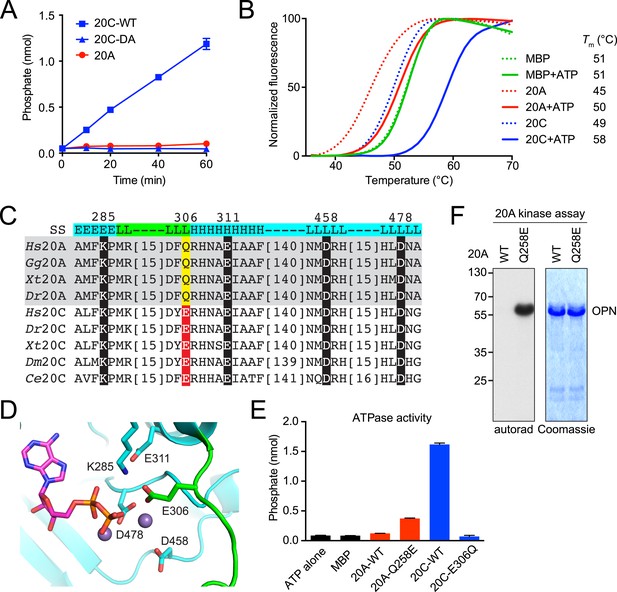
Fam20A lacks a residue critical for kinase activity.
(A) Intrinsic ATPase activity of Fam20C, Fam20C D478A (DA) and Fam20A. Recombinant proteins were incubated with 0.5 mM ATP and 10 mM MnCl2 at 30°C for the indicated time. The amount of phosphate released during incubation was determined using malachite green reagent. (B) Thermal stability shift assay of MBP, Fam20A and Fam20C. Protein thermal stability was monitored by the fluorescence generated from binding of the dye SYPRO Orange to the hydrophobic region exposed upon protein denaturation. MBP purified the same way as Fam20A and Fam20C was used as a negative control. Reaction buffer contained 10 mM MnCl2. (C) Sequence alignment of Fam20A and Fam20C protein orthologs (Hs, Homo sapiens; Gg, Gallus gallus; Xt, Xenopus tropicalis; Dr, Danio rerio; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans). Catalytically important residues are highlighted and numbered according to human Fam20C. See Figure 2—figure supplement 1 for an extended alignment. (D) Fam20C active site from PDB:4kqb bound to ADP and Mn2+ ions. Conserved canonical kinase ion pair, ion interacting, and catalytic residues are highlighted (cyan sticks) and labeled according to human Fam20C. A Fam20-specific loop is colored in green and contributes a unique active site residue E306. (E) Intrinsic ATPase activities of Fam20A Q258E and Fam20C E306Q. Reactions were carried out for 1 hr. (F) Effect of Q258E mutation on Fam20A kinase activity. Recombinant OPN was used as the substrate.
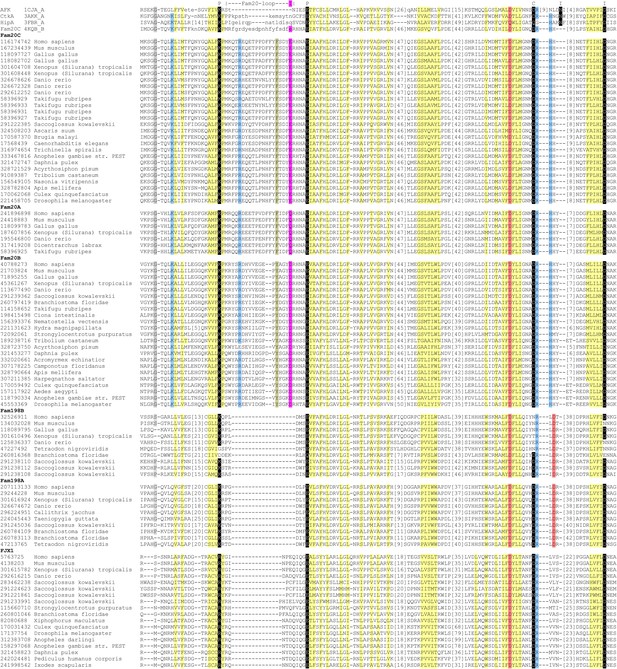
Structure-guided sequence alignment of Fam20C-related atypical kinases.
Labels above the line: P, polar salt bridge pair; I, ion coordinating; C, catalysis. Color codes: gray, mainly small residues; light yellow, mainly hydrophobic residues; light blue, mainly positive residues; light red, mainly negative residues; dark yellow, mainly aromatic residues. The NCBI gi number of each sequence is shown on the left.
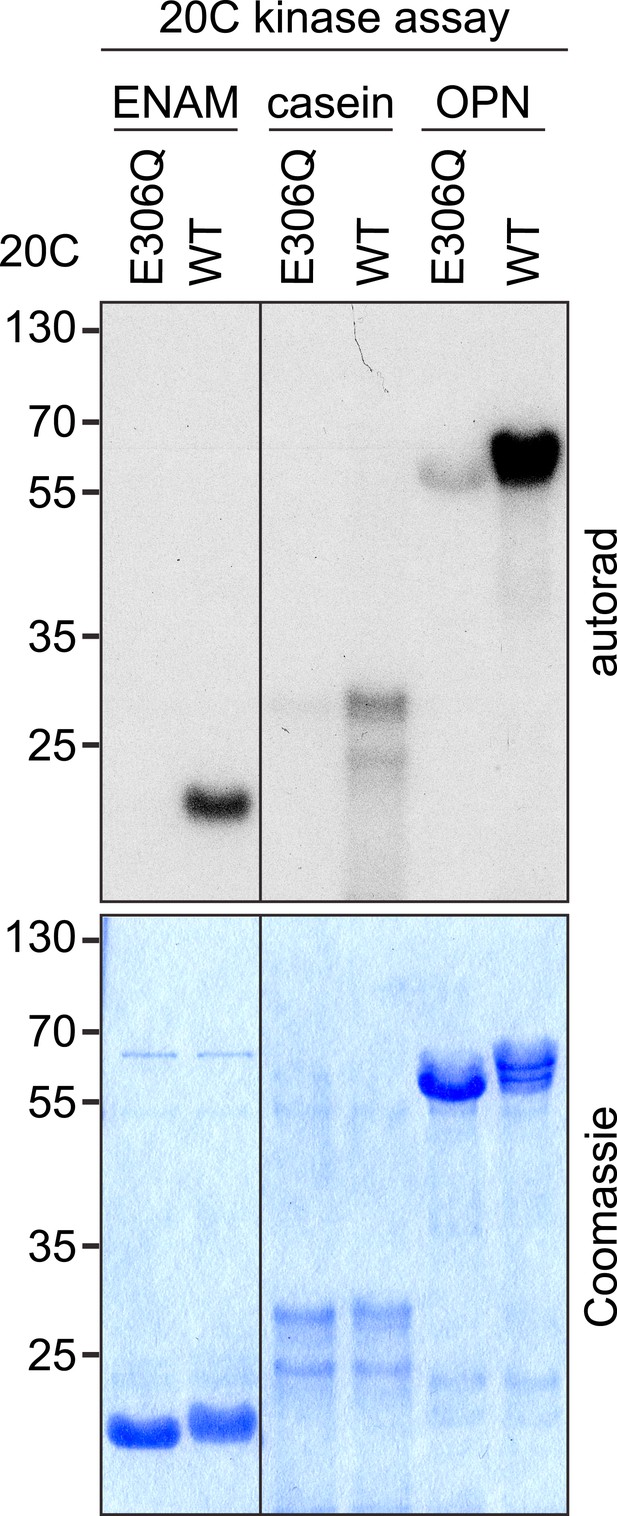
Effect of E306Q mutation on Fam20C kinase activity.
https://doi.org/10.7554/eLife.06120.007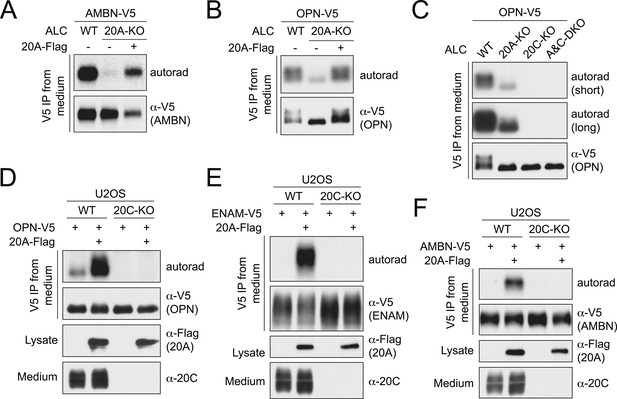
Fam20A enhances Fam20C-dependent phosphorylation.
(A and B) Effect of Fam20A KO on the phosphorylation of AMBN (A) or OPN (B). C-terminal V5-tagged AMBN or OPN (AMBN-V5 or OPN-V5) was expressed in WT or Fam20A-KO ALCs and analyzed as in Figure 1C. Flag-tagged Fam20A (20A-Flag) was co-expressed in Fam20A-KO ALCs as indicated. (C) Phosphorylation of OPN-V5 in WT, Fam20A-KO, Fam20C-KO, or Fam20A and Fam20C double-KO (A&C-DKO) ALCs. The experiment was performed as in (B). Two different exposures of autoradiography are shown. (D–F) Effect of Fam20A overexpression on the phosphorylation of OPN (D), ENAM (173–277) (E) or AMBN (F) in WT or Fam20C-KO U2OS cells. C-terminal V5-tagged OPN, ENAM (173–277) or AMBN (OPN-V5, ENAM-V5 or AMBN-V5) were expressed in WT or Fam20C-KO U2OS cells and analyzed as in Figure 1C. Fam20A-Flag was stably expressed as indicated. Fam20A-Flag in the cell lysates and endogenous Fam20C in the conditioned medium are also shown. Fam20C-KO U2OS cells were generated by means of transcription activator-like effector nuclease (TALEN; Figure 3—figure supplement 2).
CRISPR-mediated Fam20A KO and Fam20A/Fam20C double KO (DKO) in ALCs.
(A) The schematic of the sgRNA-targeting site in the mouse Fam20a gene and the disrupted allele in Fam20A-KO ALCs (clone A21). The first exon of the mouse Fam20a gene is shown as a box. White, 5′ UTR; Orange, the coding sequence. Targeting sites and PAMs are indicated with blue and red bars, respectively. Inserted nucleotides are highlighted in red. (B) Disrupted mouse Fam20c allele in Fam20A/Fam20C-DKO ALC (clone AC33). Fam20A-KO ALC clone A21 (Figure 3—figure supplement 1A) was used as the parental cell line. Fam20c gene was targeted using sgRNA m20c #3 (Figure 1—figure supplement 1A). (C) Endogenous Fam20C production by WT ALC or the KO clones used in this study. Cells were seeded at the same density and cultured overnight in serum-free media. Proteins in the conditioned media were precipitated using trichloroacetic acid (TCA) and separated by SDS-PAGE. Secreted Fam20C was detected by western blot (upper). Equal protein loading of each sample was shown by Ponceau S staining (lower).
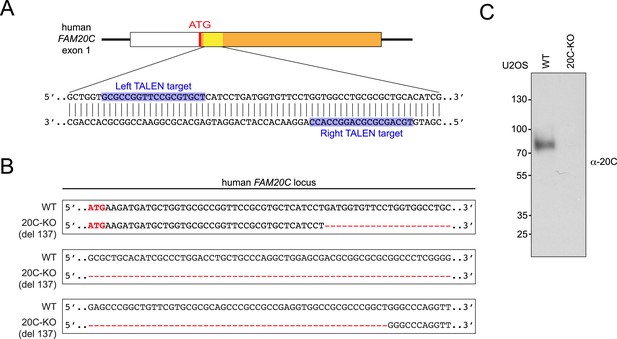
Transcription activator-like effector nuclease (TALEN)-mediated Fam20C KO in U2OS cells.
(A) The schematic of the TALEN-targeting sites in the human FAM20C gene. (B) Disrupted human FAM20C allele in Fam20C-KO U2OS cells. Deleted region is indicated with red dashes. (C) Endogenous Fam20C production by WT or Fam20C-KO U2OS. The experiment was performed as in Figure 3—figure supplement 1C.
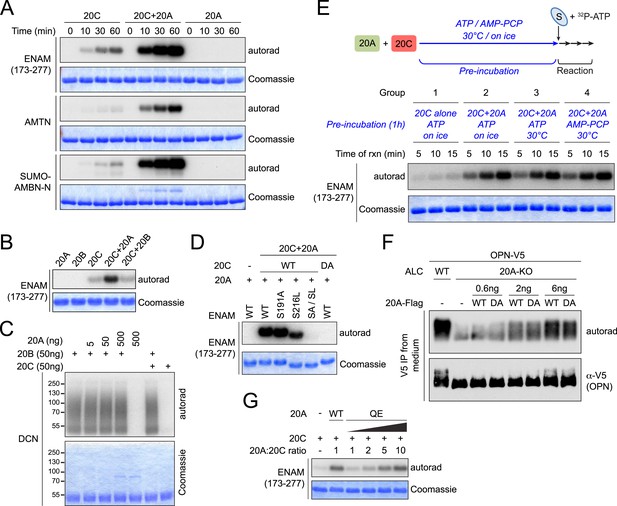
Fam20A increases Fam20C kinase activity in vitro.
(A) Time-dependent 32P incorporation into enamel matrix proteins by Fam20C, Fam20A, or the combination of Fam20A and Fam20C (molar ratio of 1:1). SUMO-AMBN-N, a N-terminal peptide of AMBN fused to a SUMO tag. (B) Effect of Fam20B on ENAM phosphorylation by Fam20C. (C) Effect of Fam20A or Fam20C on Fam20B-catalyzed phosphorylation of proteoglycan decorin. Fam20B specifically phosphorylated glycosylated decorin, which migrated as an elongated smear due to the heterogeneity of attached glycosaminoglycans (Wen et al., 2014). (D) Effect of SxE motif mutations on ENAM phosphorylation by the combination of Fam20A and Fam20C. The experiment was performed as in Figure 1B, except that equal amounts of Fam20A and Fam20C were added to the reaction. DA, inactive Fam20C D478A. (E) Effect of pre-incubation of Fam20A and Fam20C on ENAM phosphorylation. Fam20A (20 μg/ml) and Fam20C (0.5 μg/ml) were pre-incubated with ATP or nonhydrolyzable AMP-PCP (50 μM), on ice or at 30°C for 1 hr as indicated. Recombinant ENAM (173–277) and excessive ATP were then added to the final concentration of 250 μg/ml and 1 mM, respectively. [γ-32P]-ATP was added on the second step to trace ENAM phosphorylation. (F) Comparison of the abilities of Fam20A WT and D430A to restore OPN phosphorylation in Fam20A-KO ALCs. The experiment was performed as in Figure 3B. For transfection, 0.6–6 ng of 20A-Flag (as indicated) and 2 μg of OPN-V5 DNA were used for each 35 mm dish. (G) Effect of Fam20A Q258E (QE) on ENAM phosphorylation by Fam20C in vitro.
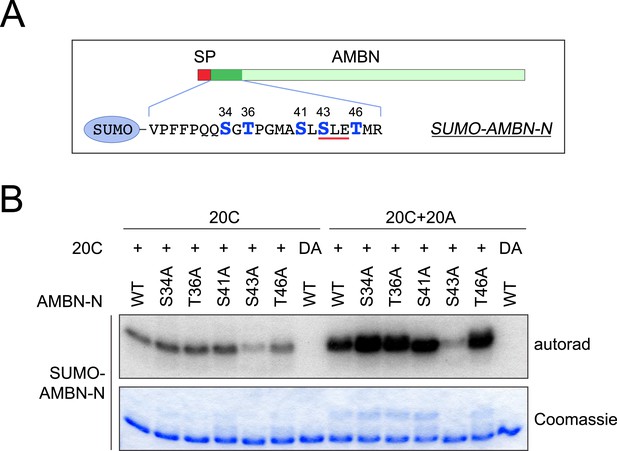
Mutagenesis scanning for the phosphorylation site of SUMO-AMBN-N.
(A) The schematic of human AMBN and the SUMO-tagged AMBN N-terminal peptide (SUMO-AMBN-N). Ser and Thr residues are highlighted and numbered. A red bar indicates the only SxE motif in SUMO-AMBN-N. SP, signal peptide. (B) SUMO-AMBN-N or its mutants were incubated with Fam20C alone or the combination of Fam20A and Fam20C (molar ratio of 1:1) in the presence of [γ-32P]-ATP as indicated. S43 within an SxE motif appeared to be the primary phosphorylation site, either in the presence or absence of Fam20A. DA, inactive Fam20C D478A.
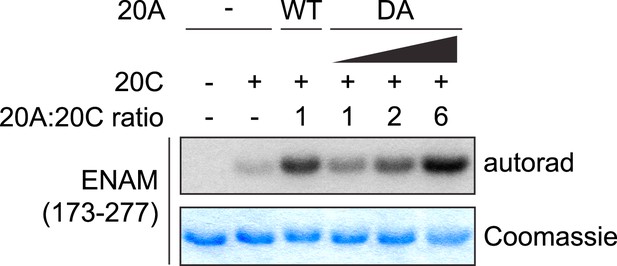
Effect of Fam20A D430A on ENAM phosphorylation by Fam20C in vitro.
Variable amount of Fam20A D430A (DA) was used. The molar ratio between Fam20A and Fam20C in each lane is indicated.
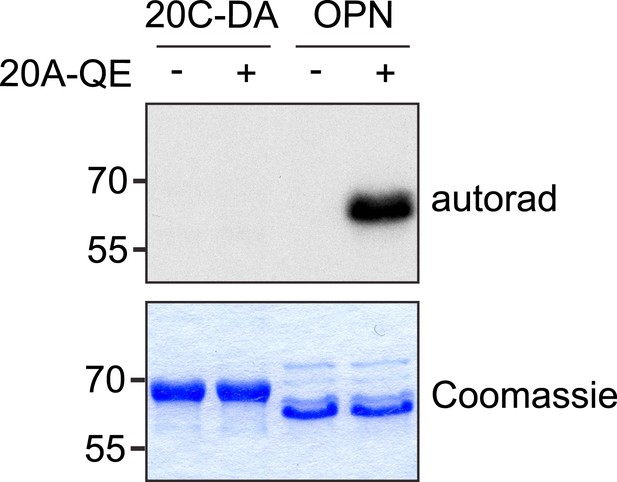
Fam20A Q258E does not phosphorylate Fam20C.
Inactive Fam20C D478A was used as the substrate to avoid Fam20C autophosphorylation. OPN was used as a positive control.
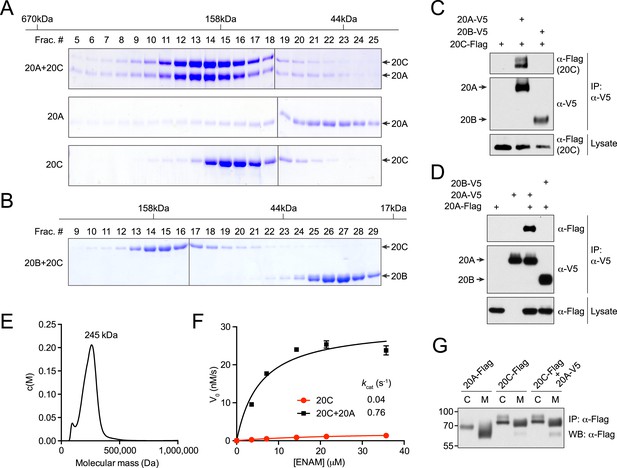
Fam20A and Fam20C form a complex that is catalytically more active.
(A) Gel filtration analysis of Fam20A/Fam20C complex. Purified Fam20A and Fam20C were pre-incubated on ice and passed through Superdex 200. Proteins from different fractions were separated by SDS-PAGE and visualized by Coomassie staining. Fam20A or Fam20C alone were analyzed as controls. The calibration standard is shown on top. (B) Gel filtration analysis of Fam20B and Fam20C. (C) Co-immunoprecipitation of Fam20A and Fam20C from cells. Fam20C-Flag, Fam20A-V5, or Fam20B-V5 was stably expressed in U2OS cells as indicated. (D) Co-immunoprecipitation of Fam20A-Flag and Fam20A-V5 from U2OS cells. (E) Analytical ultracentrifugation sedimentation velocity analysis of Fam20A/Fam20C complex. The plot represents the molecular mass distribution c(M) vs the apparent molecular mass (Da). The main peak has a calculated weight of 245 kDa. (F) Fam20C kinase reaction initial velocities vs concentration of ENAM (173–277) S191A. Fam20C (2 μg/ml) and Fam20A (40 μg/ml) were used in this experiment. Data points are represented as mean ± SD and fitted by non-linear regression of the Michaelis-Menten equation. (G) Immunoprecipitation of Fam20A-Flag or Fam20C-Flag from either cell lysate (C) or the conditioned medium (M). U2OS cells stably expressing Fam20A-Flag, Fam20C-Flag or both Fam20C-Flag and Fam20A-V5 were used.
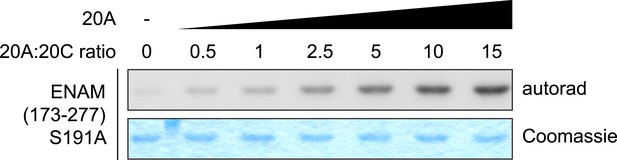
Saturation of Fam20C kinase activity by Fam20A in vitro.
Fam20C kinase reactions were performed as in Figure 5F using ENAM (173–277) S191A as the substrate. Increasing amount of Fam20A was added to the reaction. The molar ratio between Fam20A and Fam20C in each lane is indicated. Saturation of ENAM phosphorylation was observed when 10 to 15-fold excess of Fam20A were used.
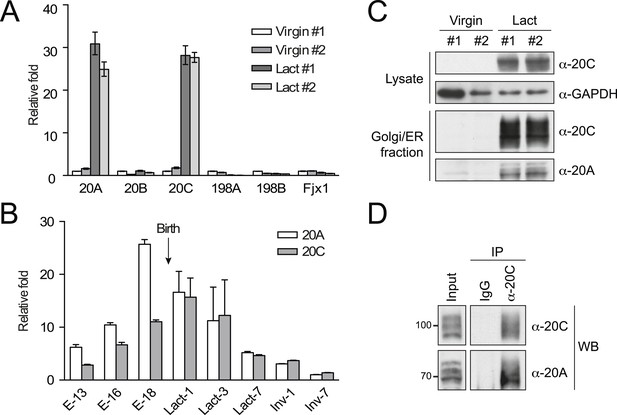
Upregulation and complex formation of Fam20A and Fam20C in the lactating mammary gland.
(A) mRNA levels of Fam20C family members in the mouse mammary gland. The mammary glands from two non-lactating (Virgin) and two 10-day lactating (Lact) mice were isolated and the mRNA levels of Fam20 family members were determined by quantitative (q)RT-PCR. Data are represented as mean ± SD. (B) mRNA levels of Fam20A and Fam20C in the mammary glands of pregnant/lactating female mice at different stages. E−13, 16 or 18: embryonic day 13, 16 or 18; Lact-1, 3 or 7: 1, 3 or 7 days after lactation; Inv-1 or 7: 1 or 7 days after involution. The gestation period of mouse is 18–21 days. mRNA levels of Fam20A and Fam20C were determined by qRT-PCR. Data are represented as mean ± SD. (C) Protein levels of Fam20A and Fam20C in the mouse mammary glands. Endogenous Fam20A or Fam20C were detected from whole mammary gland lysate or the Golgi/ER-enriched fractions by western blot. (D) Co-immunoprecipitation of endogenous Fam20A and Fam20C from Golgi/ER-enriched fractions of the mouse lactating mammary gland. Polyclonal rabbit anti-Fam20C antibody was used for immunoprecipitating endogenous Fam20C. Normal rabbit IgG was used as control for immunoprecipitation.
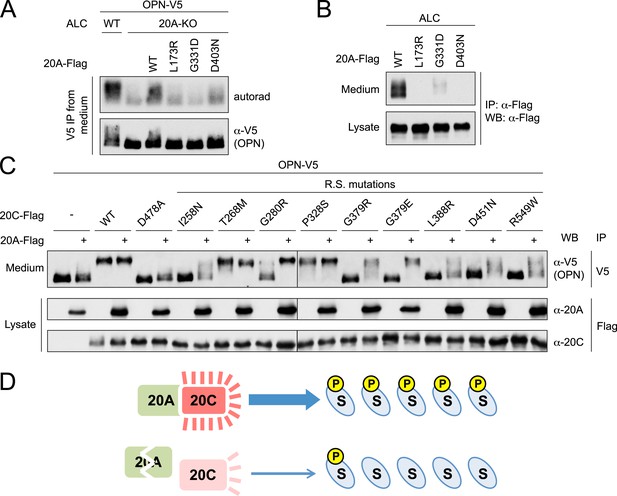
Fam20A mutants found in AI patients fail to enhance Fam20C activity, and WT Fam20A is able to increase the activities of Fam20C mutants.
(A) Effect of Fam20A mutations on OPN phosphorylation. The experiment was performed as in Figure 3B. OPN-V5 was expressed in WT or Fam20A-KO ALC. Fam20A-Flag WT, L173R, G331D or D403N was co-expressed as indicated. (B) Effect of AI mutations on Fam20A secretion. Fam20A-Flag WT, L173R, G331D or D403N was expressed in ALCs. Fam20A-Flag was immunoprecipitated from conditioned media or cell lysate using anti-Flag antibody and detected by western blot. (C) Effect of Fam20A overexpression on Fam20C mutant-catalyzed OPN phosphorylation. OPN-V5 and Fam20C-Flag (WT or mutants as indicated) were expressed in U2OS cells with or without Fam20A-Flag. Secreted OPN-V5 was immunoprecipitated from conditioned media and detected by western blot. OPN phosphorylation was indicated by its mobility shift. D478A is the kinase-dead Fam20C mutation. Other Fam20C mutations were identified from Raine syndrome (R.S.) patients. (D) The working model of Fam20A/Fam20C complex. Upper: in the tissues and cells where both Fam20A and Fam20C are expressed, Fam20C forms a complex with Fam20A and exhibits high activity, resulting in high levels of substrate (S) phosphorylation; Lower: when Fam20A is disrupted, Fam20C activity becomes low and substrate phosphorylation is ineffective, leading to diseases like AI.





