Dynamic regulation of transcription factors by nucleosome remodeling
Figures
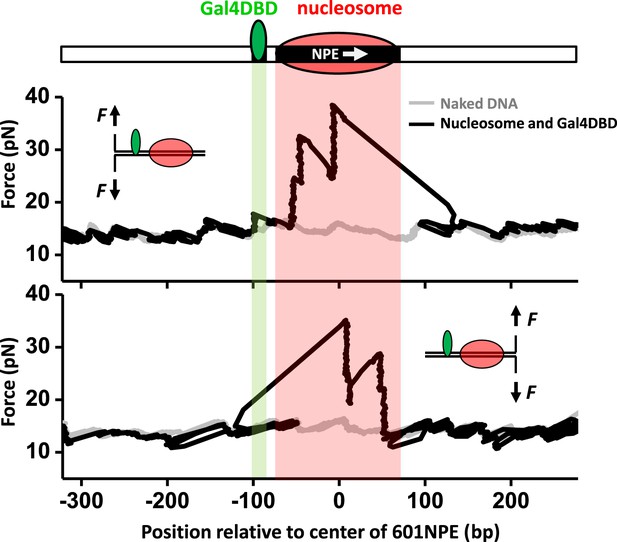
Single molecule unzipping technique detects Gal4DBD and nucleosome to near base-pair accuracy.
DNA molecules, each containing a nucleosome and a bound Gal4DBD, were unzipped. All unzipped DNA molecules used in this work were in the region of 600 bp to 1.2 kbp. For clarity, much smaller regions are shown in all figures, with the origin of a template sequence defined as center position (the dyad) of the 601NPE. Shaded regions indicate locations of the Gal4 binding sequence and the 601NPE. (top panel) Cartoon illustrating the unzipping template design using for this experiment. A Gal4 sequence was separated from a 601NPE by 10 bp. The orientation of the 601NPE sequence is indicated by a white arrow. (middle panel) Unzipping in the direction in which the bound Gal4DBD was encountered first. (bottom panel) Unzipping in the direction in which the nucleosome was encountered first.
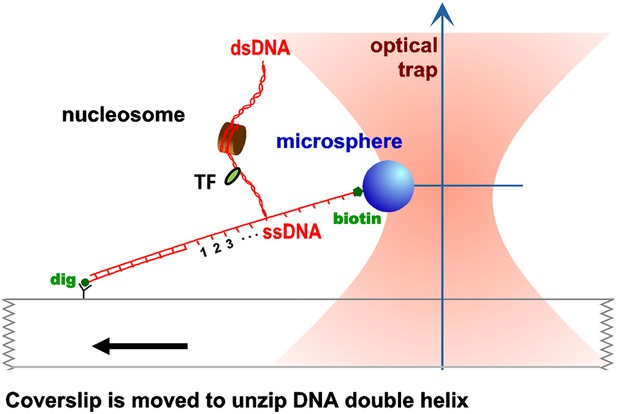
Unzipping experimental configuration.
The DNA template was attached, at one end, to the surface of a glass coverslip via a digoxigenin–antidigoxigenin linkage, and at its other end to a microsphere via a biotin–streptavidin linkage. As the coverslip was moved away from the trapped microsphere, using a loading-rate clamp, the dsDNA was sequentially converted into ssDNA upon base pair separation. The presence of force peaks above the naked DNA baseline revealed the detected locations of protein–DNA interactions.
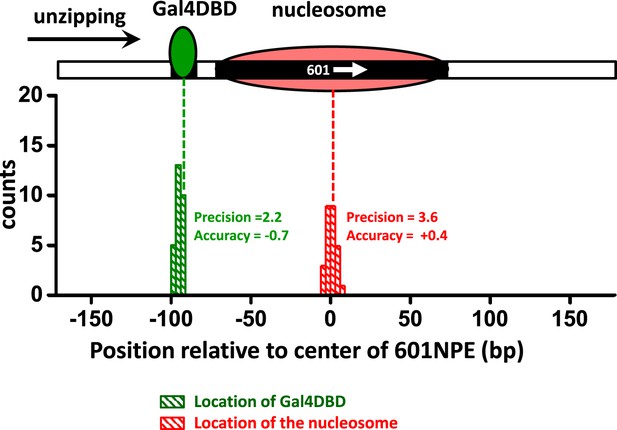
Characterization of the precision and accuracy of detection of the locations of Gal4DBD and nucleosome.
Single molecule unzipping detected Gal4DBD and a nucleosome simultaneously. The histograms for detected locations of Gal4DBD (green) and nucleosome (red) were obtained by pooling data from multiple single molecule traces, with the expected bound locations represented by their respective dashed lines. For each histogram, the precision was determined by the standard deviation of each histogram, and the accuracy by the difference between the mean of the histogram and the expected value (the vertical dashed line). These data demonstrate both the precision and accuracy to be near base-pair.
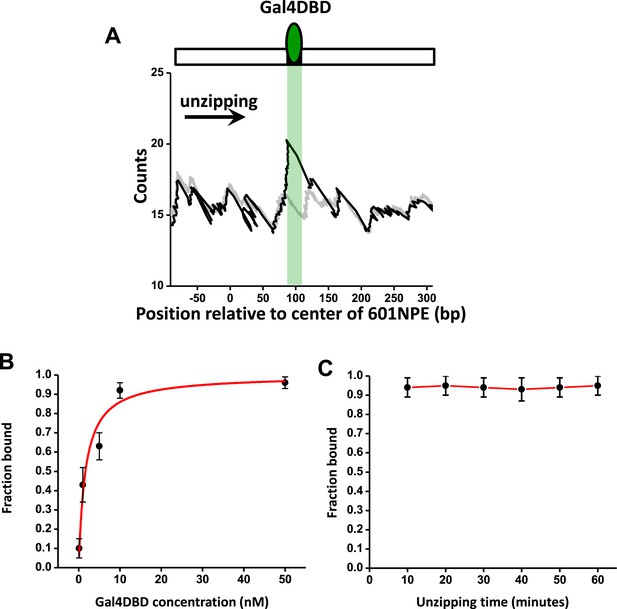
Characterization of Gal4DBD binding.
To characterize Gal4DBD binding to its binding sequence, DNA unzipping was carried out in the presence of known Gal4DBD concentrations. (A) A representative unzipping trace of a bound Gal4DBD. The location of the binding sequence is shown as a shaded region. Naked DNA unzipping baseline is shown in gray. (B) Fraction of bound Gal4DBD vs the concentration of Gal4DBD. For a given concentration of Gal4DBD, measurements were on multiple DNA molecules to obtain the fraction of Gal4DBD. Data points are represented as (mean ± s.e.m.). The relation for the fraction bound vs [Gal4DBD] was fit to: (red smooth curve), which yielded the dissociation equilibrium constant = 3.4 nM. (C) Fraction of bound Gal4DBD vs time. This relation shows no significant Gal4DBD dissociation from its binding sequence over a course of one hour. Data were fit to a straight line to guide the eye.
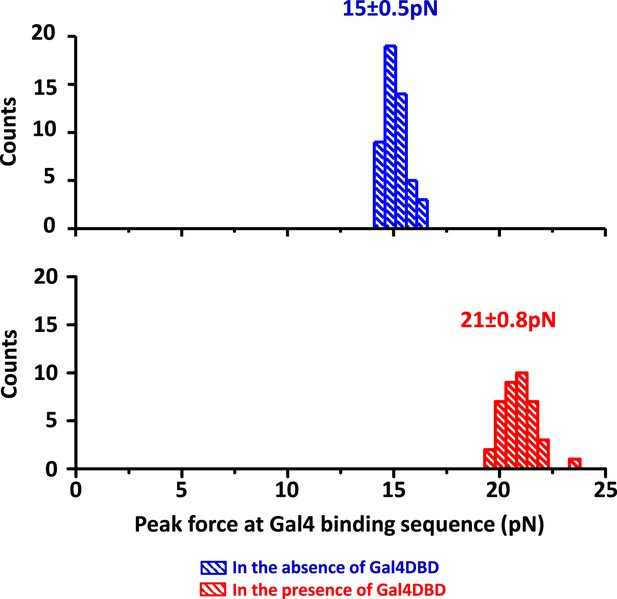
Detection of Gal4DBD binding.
The presence of a bound Gal4DBD was determined by the magnitude of the force peak at the Gal4 binding sequence. In the presence of a bound Gal4DBD, the peak force increased substantially and was readily differentiable from the baseline DNA force.
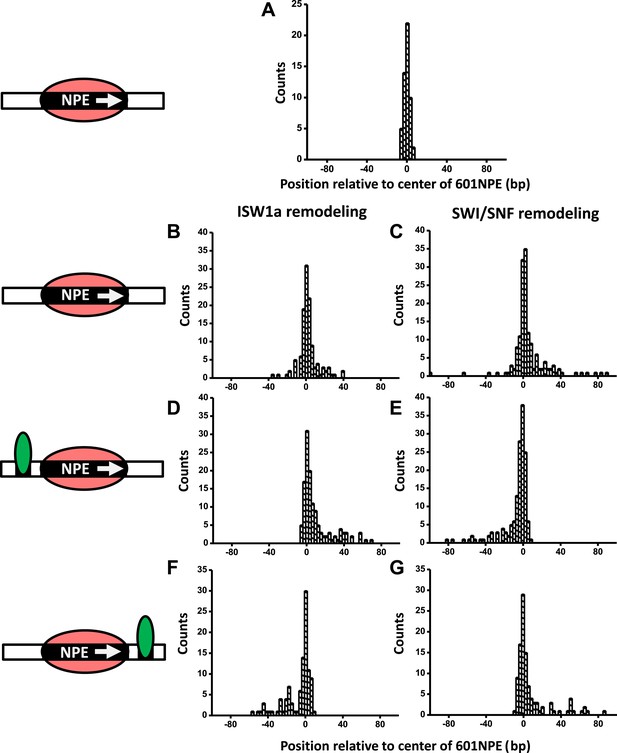
A bound Gal4DBD affects the directionality of SWI/SNF remodeling and ISW1a remodeling differently.
Nucleosomes were remodeled by either 1 nM ISW1a or 1 nM SWI/SNF with 1 mM ATP for 1 min, a time sufficiently short that the majority of nucleosomes were not remodeled (Figure 2—figure supplement 2). Each DNA template was subsequently unzipped. For templates used in (D)–(G), the 601NPE was separated from the Gal4 binding sequence by 10 bp. (A) Distribution of the location of a nucleosome before remodeling. Data were pooled from measurements on multiple nucleosomal DNA molecules. (B) Distribution of the location of a nucleosome remodeled by ISW1a in the absence of Gal4DBD. (C) Distribution of the location of a nucleosome remodeled by SWI/SNF in the absence of Gal4DBD. (D) Distribution of the location of a nucleosome remodeled by ISW1a with a bound Gal4DBD initially located upstream of the 601NPE. (E) Distribution of the location of a nucleosome remodeled by SWI/SNF with a bound Gal4DBD initially located upstream of the 601NPE. (F) Distribution of the location of the nucleosome remodeled by ISW1a with a bound Gal4DBD initially located downstream of the 601NPE. (G) Distribution of the location of a nucleosome remodeled by SWI/SNF with a bound Gal4DBD initially located downstream of the 601NPE.
-
Figure 2—source data 1
Comparison of unzipping force signatures of a nucleosome before and after remodeling.
We used unzipping to characterize the structure of a nucleosome before or after remodeling by either ISW1a or SWI/SNF, in the presence or absence of Gal4DBD. The structural features include the maximum force in the first force cluster, the maximum force in the second force cluster, the width of each cluster, and the distance between the two clusters. Errors show s.d.
- https://doi.org/10.7554/eLife.06249.009
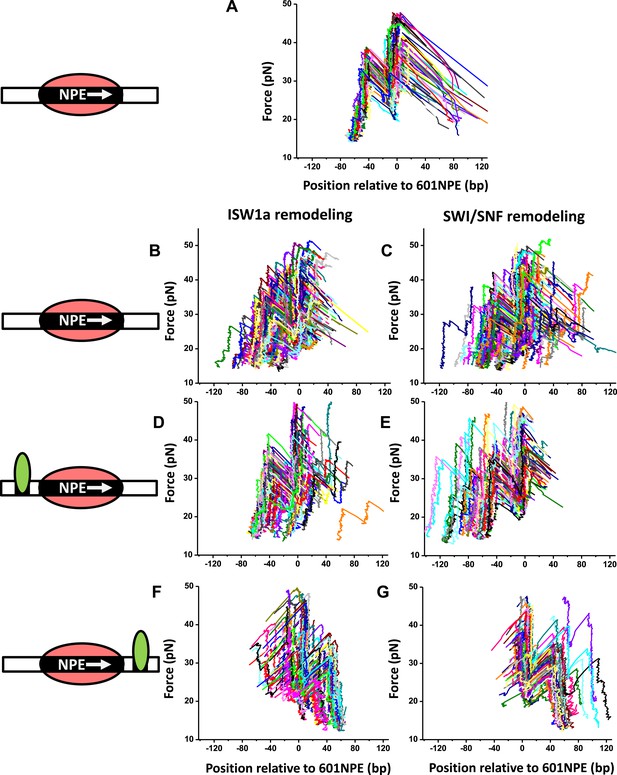
Directionality of ISW1a and SWI/SNF remodeling of a nucleosome in close proximity to a bound Gal4DBD.
The figure panels A to G show the raw traces for the corresponding panels in Figure 2.
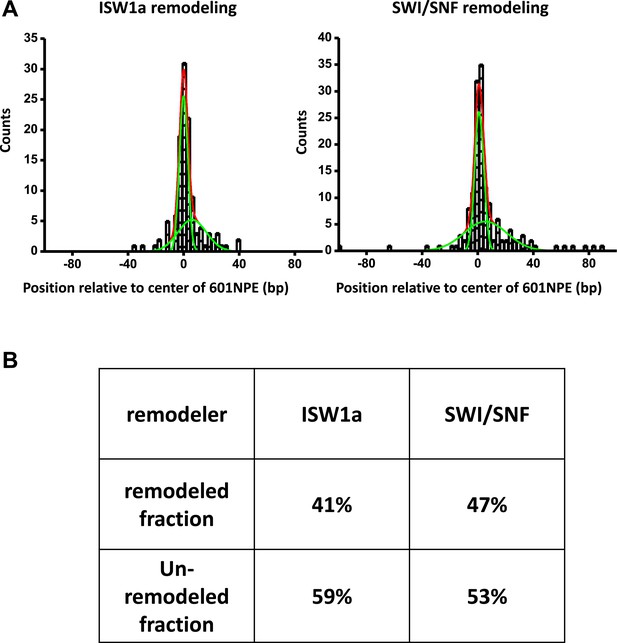
Determination of fractions of remodeled nucleosome.
(A) For each nucleosome distribution, remodeled by either ISW1a or SWI/SNF as shown in Figure 2B,C, was fit to a double Gaussian function. One Gaussian (narrow green curve) corresponds to the distribution of unremodeled nucleosomes and the other (broader green curve) to the distribution of remodeled nucleosomes. The sum of the two Gaussians is shown as the red curve. (B) A table summarizing fractions of unremodeled percentage and remodeled percentage for both ISW1a and SWI/SNF.
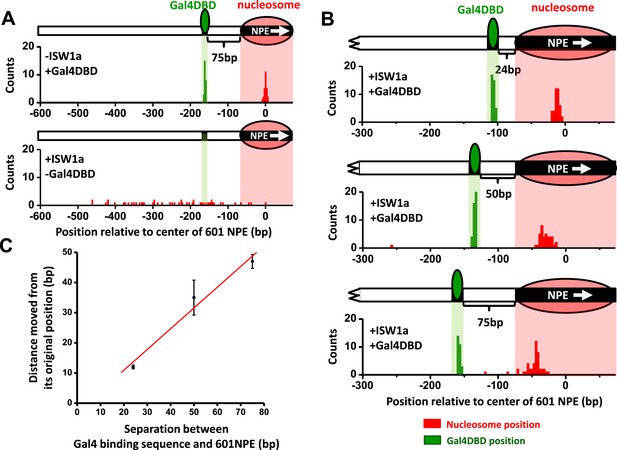
ISW1a remodeling is blocked by a bound Gal4DBD.
Nucleosomes were remodeled by 1 nM ISW1a with 1 mM ATP for 10 min with or without Gal4DBD. Shaded regions indicate locations of Gal4 binding sequence and 601NPE. (A) Distributions of the locations of the nucleosome and bound Gal4DBD, either before remodeling or without Gal4DBD, as controls. (B) Distributions of the locations of the nucleosome after ISW1a remodeling in the presence of Gal4DBD on three different templates of increasing separation between the Gal4 binding sequence and the 601NPE. For each template, the nucleosome position distribution is dominated by a narrow population, but has a few outliers which were moved a much greater distance and some of which even passed the Gal4 binding sequence. These outliers (∼5%) were likely a result of templates that did not have a bound Gal4DBD initially (∼5%; see main text). This is further supported by the observation that none of these outlier traces revealed a bound Gal4DBD. Nonetheless, in order to avoid possible bias, these nucleosome positions were still used for further analysis in (C) and thus contributed to the error bars in (C). (C) Relationship between the distance the remodeled nucleosome moved and the separation between the Gal4 binding sequence and the 601NPE. Error bars are SEM.
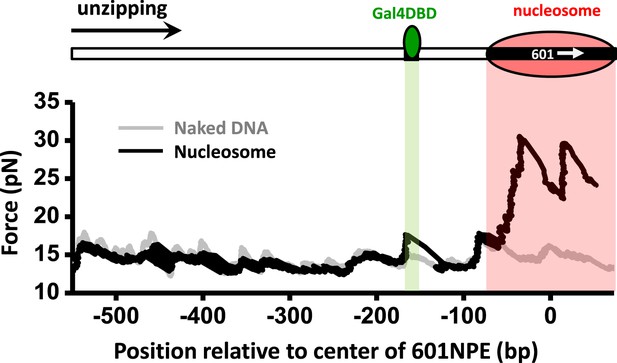
Single molecule unzipping simultaneously detects Gal4DBD and an end-positioned nucleosome.
Shown is a representative unzipping trace of a DNA molecule containing an end-positioned nucleosome and a bound Gal4DBD.
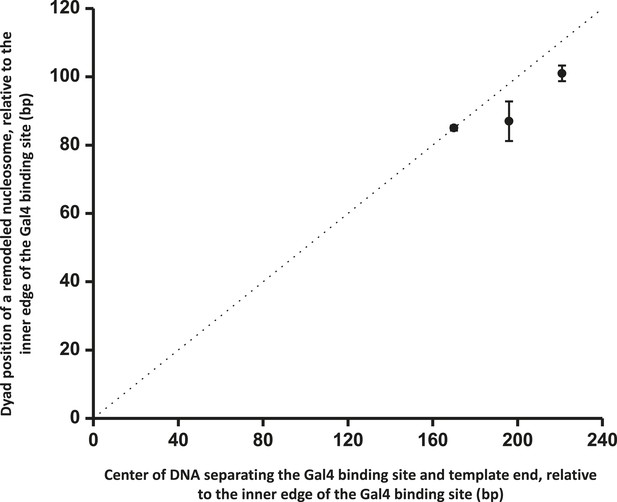
ISW1a centers a nucleosome between a bound Gal4DBD and the template end.
This relation shows that ISW1a remodeler tends to center a nucleosome between the Gal4 binding site and the DNA end. Error bars are SEM. The dashed line indicates the center position on the DNA.
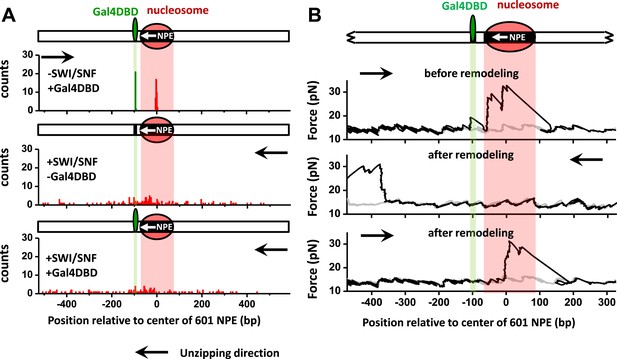
SWI/SNF remodeling evicts a bound Gal4DBD from its DNA template.
Nucleosomes were remodeled by 1.5 nM SWI/SNF with 1 mM ATP for 10 min with or without Gal4DBD. Shaded regions indicate locations of Gal4 binding sequence and 601NPE. (A) Distributions of the locations of the nucleosome and bound Gal4DBD before remodeling (upper plot), after remodeling without Gal4DBD (middle plot), and after remodeling with Gal4DBD (lower plot). (B) Representative traces in the case of before remodeling (top plot; N = 55) and after remodeling (middle and bottom plots; N = 50). The middle plot shows an example trace where a nucleosome was remodeled to the opposite side of Gal4DBD relative to its original position; while the bottom plot shows an example trace where a nucleosome was remodeled to the same side of Gal4DBD relative to its original position. Gray traces were taken from the corresponding naked DNA.
-
Figure 4—source data 1
SWI/SNF is unable to evict a bound Gal4DBD in the absence of a nucleosome or in the absence of ATP.
To determine whether SWI/SNF with ATP alone is able to displace a bound Gal4DBD in the absence of a nucleosome, we carried out experiments on a DNA template preloaded with Gal4DBD but without a nucleosome in 1.5 nM SWI/SNF with 1 mM ATP for 10 min. DNA molecules were subsequently unzipped to determine the presence of Gal4DBD. The fraction of templates containing a bound Gal4DBD remained the same before and after the remodeling reaction, indicating that SWI/SNF with ATP alone is not able to displace a bound Gal4DBD. To rule out the possibility that Gal4DBD disruption was due to binding of SWI/SNF to DNA or the nucleosome and not due to nucleosome remodeling, we carried out a control experiment on a DNA template containing a bound Gal4DBD and a nucleosome by incubating the sample with 1.5 nM SWI/SNF for 10 min in the absence of ATP. We subsequently unzipped the DNA template to determine if Gal4DBD was still bound. The fraction of templates containing a bound Gal4DBD was comparable to that of a template without a nucleosome and without SWI/SNF and ATP added, indicating that in the absence of ATP, SWI/SNF is unable to evict a bound Gal4DBD even in the presence of a nucleosome adjacent to a bound Gal4DBD.
- https://doi.org/10.7554/eLife.06249.016
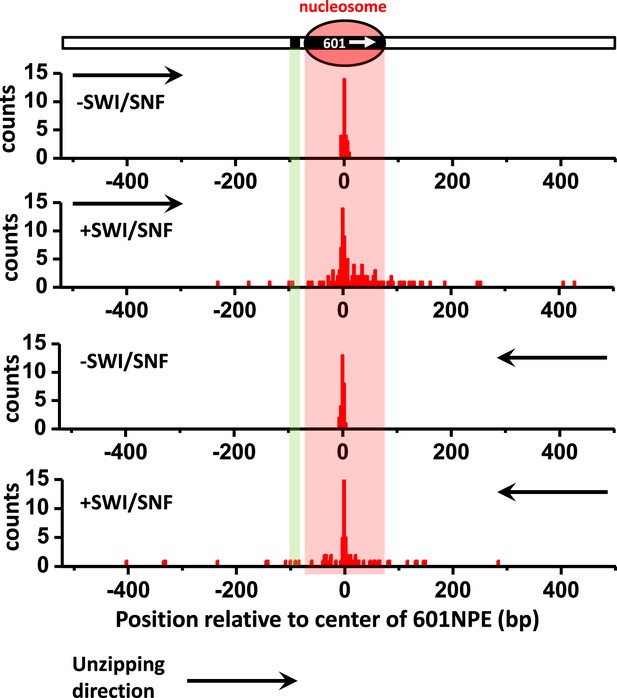
Distributions of the locations of SWI/SNF remodeled nucleosomes as determined by unzipping from both directions.
To determine whether the distributions of the locations of remodeled nucleosomes were similar for measurements made by unzipping the DNA in one direction vs in the other direction, we unzipped multiple DNA molecules, each containing either an unremodeled or a remodeled nucleosome, from both directions. Nucleosome remodeling was carried out in 1 nM SWI/SNF with 1 mM ATP for 5 min. Our data show similar distributions for data obtained in both directions.
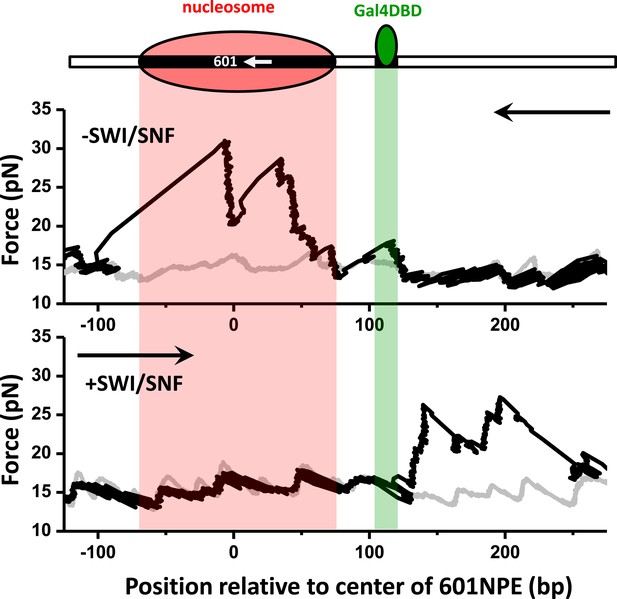
Nucleosome remodeling by SWI/SNF on a template with the Gal4 binding site separated from the 601NPE by 24 bp.
To test whether SWI/SNF is able to evict Gal4DBD via nucleosome remodeling when a bound Gal4DBD is located farther away from a nucleosome, we used a template where the Gal4 binding site was separated from the 601NPS by 24 bp and carried out unzipping experiments under identical conditions as those shown in Figure 4. Out of all traces where the nucleosome was repositioned to the opposite side of the Gal4 binding site by SWI/SNF (N = 13), we did not detect any Gal4DBD binding signature on the template, indicating eviction of Gal4DBD. Shown are example traces, with arrows indicating the unzipping directions.
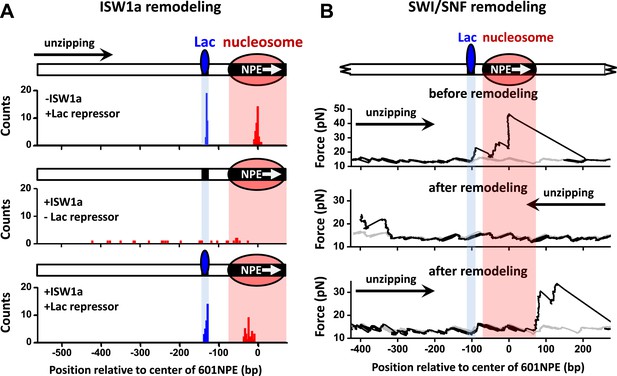
ISW1a remodeling is blocked by Lac repressor, while SWI/SNF remodeling evicts Lac repressor from the template.
Shaded regions indicate locations of Lac repressor binding sequence and 601NPE. (A) Distributions of locations of nucleosomes before remodeling (upper plot), after remodeling by ISW1a without Lac repressor (middle plot), and after remodeling by ISW1a with Lac repressor (lower plot). Lac repressor binding sequence was separated from the 601NPE by 50 bp. Nucleosomes remodeling was carried out in 1 nM ISW1a with 1 mM ATP for 10 min with or without Lac repressor. (B) Representative traces in the case of before SWI/SNF remodeling (top plot; N = 25) and after remodeling (middle and bottom plots; N = 27). Lac repressor binding site was separated from the 601NPE by 10 bp. Nucleosomes were remodeled by 1.5 nM SWI/SNF with 1 mM ATP for 10 min. The middle plot shows an example trace where a nucleosome was remodeled to the other side of the Lac repressor and the bottom plot shows an example trace where a nucleosome was remodeled to the same side of Lac repressor.
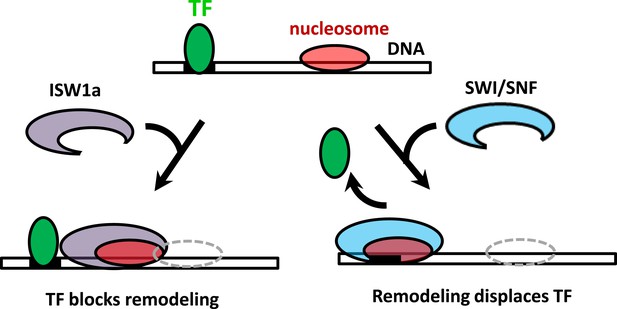
The interplay between nucleosome remodeling and a bound TF.
When a nucleosome is remodeled by ISW1a (left), nucleosome repositioning is blocked by a bound TF, the TF remains intact, and the remodeled nucleosome is positioned with the TF acting as a boundary and reference point. On the other hand, when a nucleosome is remodeled by SWI/SNF (right), nucleosome positioning is unaffected by a bound TF and the TF is evicted.





