Lpcat3-dependent production of arachidonoyl phospholipids is a key determinant of triglyceride secretion
Figures
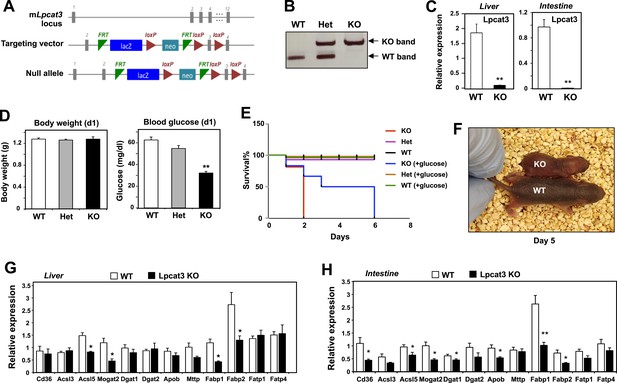
Generation and analysis of global Lpcat3 knockout mice.
(A) Strategy for generating global Lpcat3 knockout mice. A ‘knock-out first/conditional-ready’ gene-targeting vector was used to generate targeted cells. A gene-trap cassette is located between the two FRT sites. LacZ, β-galactosidase; neo, neomycin phosphotransferase II. (B) Genotyping of Lpcat3+/+ (WT), Lpcat3+/− (Het), and Lpcat3−/− (KO) mice. Genomic DNA was prepared from tail biopsies, and PCR products were separated on a 1% agarose gel. (C) Expression of Lpcat3 in liver and small intestine of newborn Lpcat3−/− and Lpcat3+/+ pups. Gene expression was quantified by real-time PCR (n ≥ 5/group). Values are means ± SEM. (D) The body weight and blood glucose of Lpcat3+/+ (WT), Lpcat3−/+ (Het), and Lpcat3−/− (KO) newborn pups (n ≥ 8/group). Values are means ± SEM. (E) Kaplan–Meier survival curve of Lpcat3+/+ (WT), Lpcat3−/+ (Het) and Lpcat3−/− (KO) pups after birth (n ≥ 20 mice/group). The neonatal lethality can be delayed by injection of 50 μl 10% glucose solution once per day after born. 5 Lpcat3+/+ (WT), 9 Lpcat3−/+ (Het) and 6 Lpcat3−/− (KO) mice were used in the rescue experiment. (F) Representative photograph of Lpcat3+/+ (WT) and Lpcat3−/− (KO) pups after 5 days of glucose injections. (G–H) Gene expression in livers (G) and small intestines (H) of Lpcat3+/+ (WT) and Lpcat3−/− (Lpcat3 KO) newborn pups. Gene expression was quantified by real-time PCR (n ≥ 5/group). Values are means ± SEM. Statistical analysis was performed using Student's t-test (C, G and H) and one-way ANOVA with Bonferroni post-hoc tests (D). *p < 0.05; **p < 0.01.
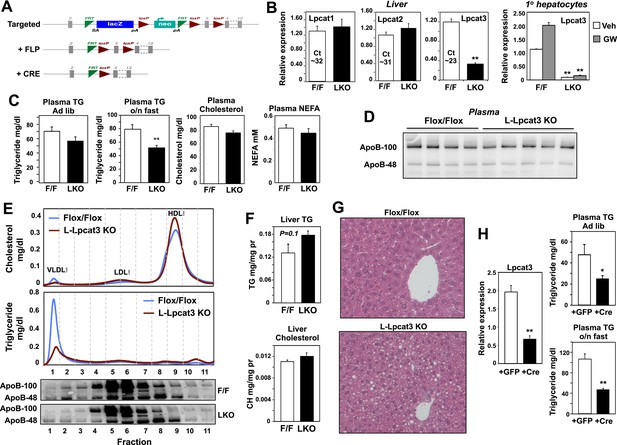
Altered triglyceride (TG) metabolism in liver-specific Lpcat3 knockout mice.
(A) Strategy for generating tissue-specific Lpcat knockout mice. Lpcat3−/+ mice carrying the conditional-ready knockout allele were mated with Flpe transgenic mice to generate the Lpcat3fl/fl mice. Lpcat3fl/fl mice were bred with tissue-specific Cre transgenic mice to generate tissue-specific Lpcat3 knockout mice. (B) Expression of Lpcat family members in liver of Lpcat3fl/fl (F/F) and Lpcat3fl/fl Albumin-Cre (L-KO) mice fed a chow diet. Gene expression was quantified by real-time PCR (n ≥ 6/group) (left panel). Primary hepatocytes from Lpcat3fl/fl (F/F) and Lpcat3fl/fl Albumin-Cre (L-KO) mice were treated overnight with 1 μM LXR agonist GW3965 (GW). Lpcat3 expression was quantified by real-time PCR (right panel). Ct values in Lpcat3fl/fl liver samples were shown. Values are means ± SEM. (C) Plasma lipid levels in chow diet-fed Lpcat3fl/fl (F/F) and Lpcat3fl/fl Albumin-Cre (L-KO) mice under fed (ad libitum) or overnight (o/n) fasting (n ≥ 8/group). Plasma levels of cholesterol and NEFA were measured after an overnight fast. Values are means ± SEM. (D) Plasma was harvested from chow diet-fed Lpcat3fl/fl (F/F) and Lpcat3fl/fl Albumin-Cre (L-KO) mice after an overnight fast. ApoB protein was analyzed by western blotting and quantified (Figure 2—figure supplement 2B). (E) Plasma from Lpcat3fl/fl (F/F) and Lpcat3fl/fl Albumin-Cre (L-Lpcat3-KO) mice fasted overnight was pooled (n = 5). Lipoprotein profiles were analyzed by fast protein liquid chromatography (FPLC) (upper panel). ApoB protein in each corresponding fraction was analyzed by western blot (lower panel) and quantified (Figure 2—figure supplement 2A). (F) Lipid contents in livers of chow diet-fed Lpcat3fl/fl (F/F) and Lpcat3fl/fl Albumin-Cre (L-KO) mice fasted overnight (n ≥ 8/group). Values are means ± SEM. (G) Hematoxylin and eosin staining of liver sections from chow-fed Lpcat3fl/fl (F/F) and Lpcat3fl/fl Albumin-Cre (L-KO) mice after an overnight fast. (H) Liver expression of Lpcat3 (left panel) and plasma TG levels (right panel) in Lpcat3fl/fl mice 3 weeks after being injected with an adenovirus encoding GFP or Cre. Mice were sacrificed under fed conditions (ad libitum) or after overnight fasting (o/n) (n ≥ 6/group). Gene expression was measured by real-time. Values are means ± SEM. Statistical analysis was performed with a Student's t-test (B, C, F, H). *p < 0.05; **p < 0.01.
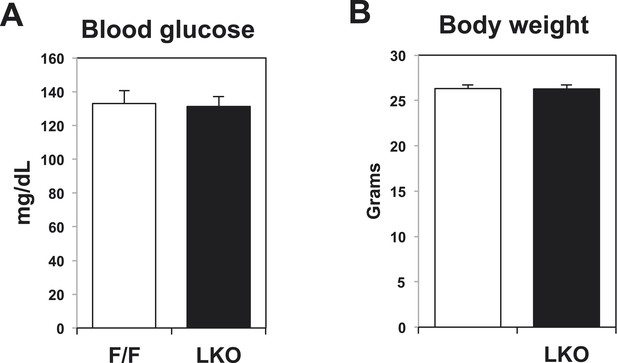
Blood glucose levels and body weight in control and liver-specific Lpcat3 KO mice (LKO).
https://doi.org/10.7554/eLife.06557.006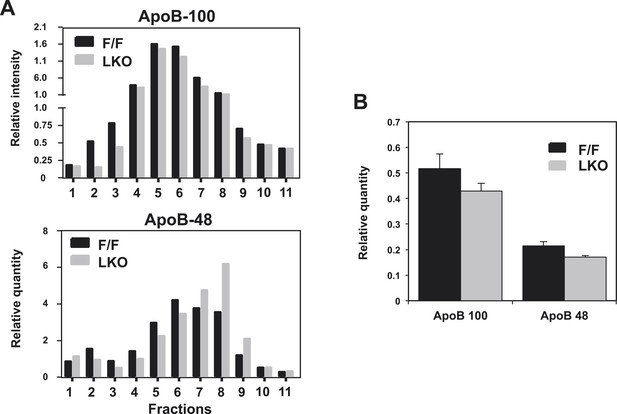
Quantification of western blots.
Immunoblot results of apoB-100 and apoB-48 proteins in (A) Figure 2E and (B) Figure 2D were quantified by densitometry.
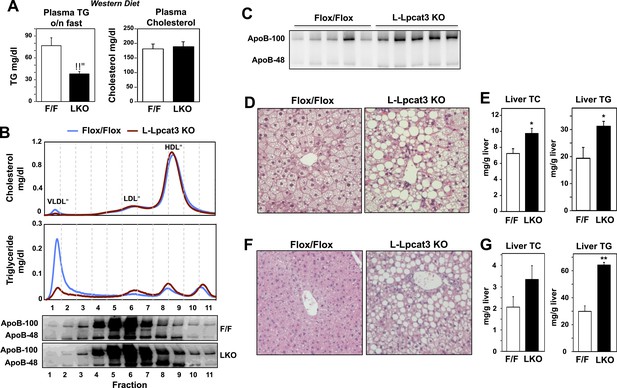
Dietary challenge accentuates metabolic phenotypes in liver-specific Lpcat3 knockout mice.
(A) Plasma lipids of Lpcat3fl/fl (F/F) and Lpcat3fl/fl Albumin-Cre (L-KO) mice fed on a western diet for 9 weeks. Plasma was collected from mice fasted for 6 hr (n ≥ 8/group). Values are means ± SEM. (B) Plasma samples same as in (A) were pooled (n = 5). Lipoprotein profile was analyzed by FPLC (upper panel). ApoB protein in each fraction was analyzed by western blot (lower panel) and quantified (Figure 3—figure supplement 1A). (C) ApoB protein in plasma samples same as in (A) was analyzed by western blot and quantified (Figure 3—figure supplement 1B). (D–E) Hematoxylin and eosin staining (D) and lipid contents (E) of livers from western diet-fed Lpcat3fl/fl (F/F) and Lpcat3fl/fl Albumin-Cre (L-KO) mice. Values are means ± SEM. (F–G) Hematoxylin and eosin staining (F) and lipid contents (G) of livers from high sucrose diet-fed Lpcat3fl/fl (F/F) and Lpcat3fl/fl Albumin-Cre (L-KO) mice. Mice were fed on diet for 3 weeks and sacrificed after 6 hr fasting. Values are means ± SEM. Statistical analysis was performed using Student's t-test (A, E and G). *p < 0.05; **p < 0.01.
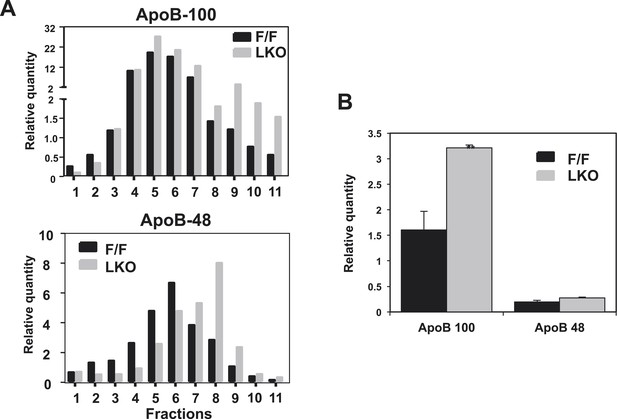
Quantification of western blots.
Immunoblot blot results of apoB-100 and apoB-48 proteins in (A) Figure 3B and (B) Figure 3C were quantified by densitometry.
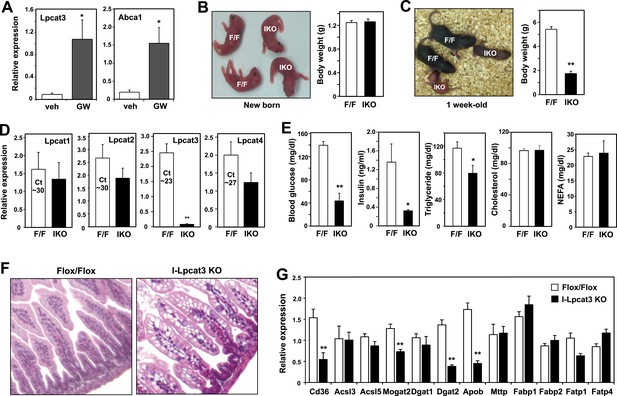
Altered TG metabolism in intestine-specific Lpcat3 knockout mice.
(A) Induction of Lpcat3 mRNA expression in duodenum of mice treated with 40 mg/kg/day GW3956 by oral gavage for 3 days (n = 5/group). Gene expression was measured by real-time PCR. Values are means ± SEM. (B) Representative photograph and body weight of newborn Lpcat3fl/fl Villin-cre (IKO) and control Lpcat3fl/fl (F/F) pups (n = 5/group for body weight measurement). Values are means ± SEM. (C) Representative photograph and body weight of 1 week-old Lpcat3fl/fl Villin-cre (IKO) and control Lpcat3fl/fl (F/F) pups (n ≥ 6/group for body weight measurement). Values are means ± SEM. (D) Expression of Lpcat family members in 1 week-old Lpcat3fl/fl (F/F) and Lpcat3fl/fl Villin-cre (IKO) duodenum measured by real-time PCR (n ≥ 6/group). Ct values of F/F samples were shown. Values are means ± SEM. (E) Blood glucose, plasma lipids and insulin levels in 1 week-old Lpcat3fl/fl (F/F) and Lpcat3fl/fl Villin-cre (IKO) pups (n ≥ 6/group). Values are means ± SEM. (F) Hematoxylin and eosin staining of intestines from 1 week-old Lpcat3fl/fl (WT) and Lpcat3fl/fl Villin-cre (IKO) pups. (G) Expression of genes in duodenum of 1 week-old Lpcat3fl/fl (WT) and Lpcat3fl/fl Villin-cre (IKO) pups. Gene expression was measured by real-time PCR (n ≥ 6/group). Values are means ± SEM. Statistical analysis was performed using Student's t-test (A, B, D, E and F). *p < 0.05; **p < 0.01.
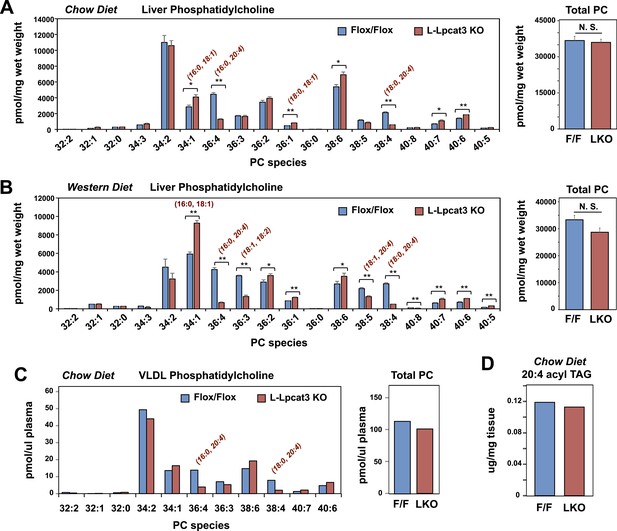
Lpcat3 is required for the incorporation of arachidonate into phosphatidylcholine (PC) in mouse liver and very low-density lipoprotein (VLDL).
(A–B) ESI-MS/MS analysis of the abundance of PC species in livers from Lpcat3fl/fl (Flox/Flox) and Lpcat3fl/fl Albumin-Cre (L-Lpcat3 KO) mice fed on a chow diet (A) and a western diet (B) (n ≥ 5/group). (C) ESI-MS/MS analysis of the abundance of PC species in plasma VLDL fraction from Lpcat3fl/fl (fl/fl) and Lpcat3fl/fl Albumin-Cre (L-Lpcat3 KO) mice fed on a chow diet. Plasma was harvested from mice after overnight fasting. VLDL fractions were pooled from 5 mice/group. (D) GC-FID analysis of the abundance of arachidonoyl acyl chain in triglyceride (TAG) in livers from Lpcat3fl/fl (F/F) and Lpcat3fl/fl Albumin-Cre (L-KO) mice fed on a chow diet. Statistical analysis was performed using Student's t-test. Values are means ± SEM. *p < 0.05; **p < 0.01.
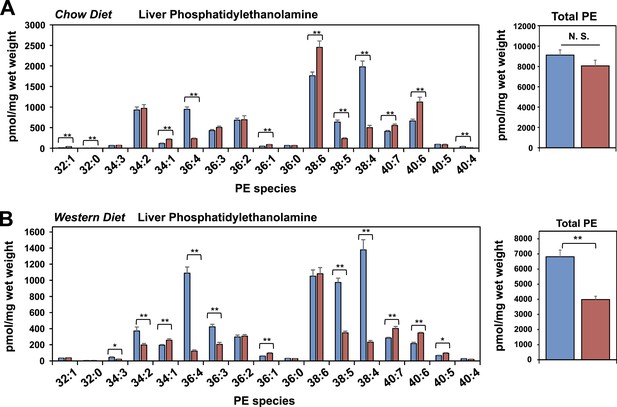
Lpcat3 is required for the incorporation of arachidonate into phosphatidylethanolamine in mouse liver.
(A–B) ESI-MS/MS analysis of the abundance of phosphatidlyethanolamine (PE) species in livers from Lpcat3fl/fl (Flox/Flox) and Lpcat3fl/fl Albumin-Cre (L-Lpcat3 KO) mice fed on a chow diet (A) and a western diet (B).
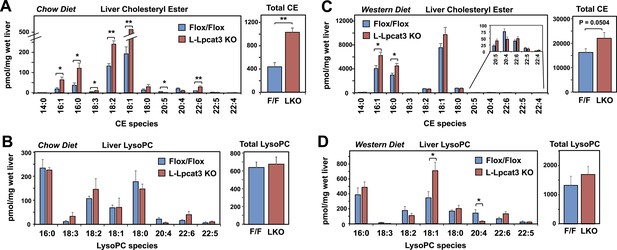
Analysis of cholesteryl ester and lysoPC species in L-Lpcat3 KO mice.
(A–B) ESI-MS/MS analysis of the abundance of cholesteryl ester (A) and lysophosphatidylcholine (LysoPC) (B) species in livers of Lpcat3fl/fl (Flox/Flox) and Lpcat3fl/fl Albumin-Cre (L-Lpcat3 KO) mice fed on a chow diet (n ≥ 5/group). (C–D) ESI-MS/MS analysis of the abundance of cholesteryl ester (C) and lysophosphatidylcholine (LysoPC) (D) species in livers of Lpcat3fl/fl (Flox/Flox) and Lpcat3fl/fl Albumin-Cre (L-Lpcat3 KO) mice fed on a western diet (n ≥ 5/group). Statistical analysis was performed using Student's t-test. Values are means ± SEM. *p < 0.05; **p < 0.01.
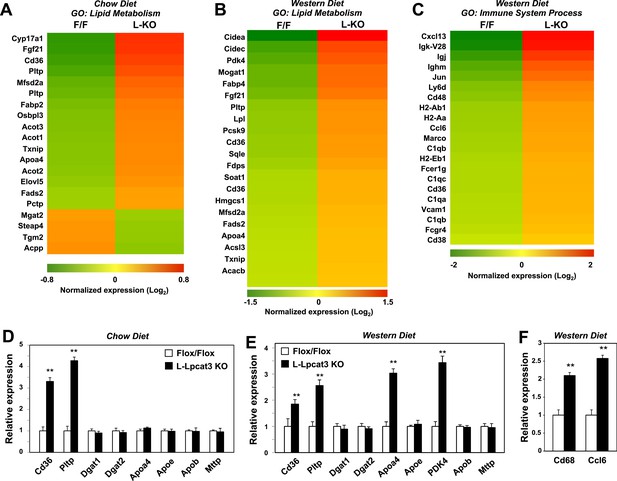
Altered gene expression linked to lipid metabolism and inflammation in liver-specific Lpcat3 knockout mice.
(A) Gene expression in livers of Lpcat3fl/fl (F/F) and Lpcat3fl/fl Albumin-Cre (L-KO) mice fed on a chow diet was analyzed by Affymetrix arrays. Select genes under the gene ontology (GO) term ‘lipid metabolism’ are presented by heatmap. Samples from 5 mice/group were pool for analysis. (B–C) Gene expression in livers of Lpcat3fl/fl (F/F) and Lpcat3fl/fl Albumin-Cre (L-KO) mice fed on a western diet for 9 weeks was analyzed by Affymetrix arrays. Select genes under the GO term ‘lipid metabolism’ (B) and ‘immune system process’ (C) are presented by heatmap. Samples from 5 mice/group were pool for analysis. (D) Expression of selective lipid metabolism genes in livers of Lpcat3fl/fl (Fl/Fl) and Lpcat3fl/fl Albumin-Cre (L-Lpcat3 KO) mice fed on a chow diet was analyzed by real-time PCR (n ≥ 5/group). Values are means ± SEM. (E–F) Expression of selective lipid metabolism (E) and inflammation (F) genes in livers of Lpcat3fl/fl (Fl/Fl) and Lpcat3fl/fl Albumin-Cre (L-Lpcat3 KO) mice fed on a western diet was analyzed by real-time PCR (n ≥ 5/group). Values are means ± SEM. Statistical analysis was performed using Student's t-test (D, E and F). **p < 0.01.
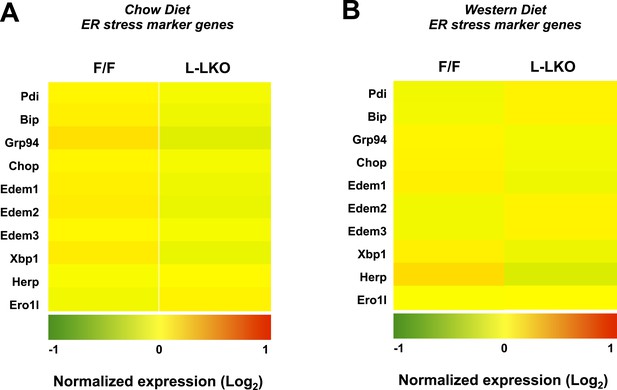
Chronic deletion of Lpcat3 does not induce unfolded protein response downstream gene expression.
(A) Gene expression in livers of Lpcat3fl/fl (F/F) and Lpcat3fl/fl Albumin-Cre (L-KO) mice fed on a chow diet was analyzed by Affymetrix arrays. Select ER stress marker genes are presented by heatmap. Samples from 5 mice/group were pool for analysis. (B) Gene expression in livers of Lpcat3fl/fl (F/F) and Lpcat3fl/fl Albumin-Cre (L-KO) mice fed on a western diet was analyzed by Affymetrix arrays. Select ER stress marker genes are presented by heatmap. Samples from 5 mice/group were pool for analysis.
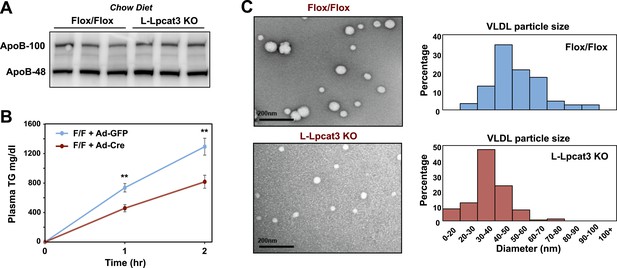
Loss of Lpcat3 from liver impairs hepatic TG secretion.
(A) ApoB protein in livers from Lpcat3fl/fl (Flox/Flox) and Lpcat3fl/fl Albumin-Cre (L-Lpcat3 KO) mice fed on a chow diet was analyzed by western blot (n = 3). (B) VLDL-TG secretion in Lpcat3fl/fl mice transduced with adenoviral expressed Cre (Ad-Cre), compared to control GPF (Ad-GFP) for 4 weeks. Mice were fasted for 6 hr followed by intravenous injection of tyloxapol. Plasma TG was measured at indicated durations after tyloxapol injection. Values are means ± SEM. (C) Plasma VLDL particle size in Lpcat3fl/fl Albumin-Cre mice. Isolated VLDL particles from Lpcat3fl/fl (Fl/Fl) and Lpcat3fl/fl Albumin-Cre (L-Lpcat3 KO) mice fed on a western diet were stained with 2.0% uranyl acetate and visualized by electron microscopy (EM) (left) and their size quantified (right). Statistical analysis was performed using two-way ANOVA with Bonferroni post hoc tests (B). **p < 0.01.
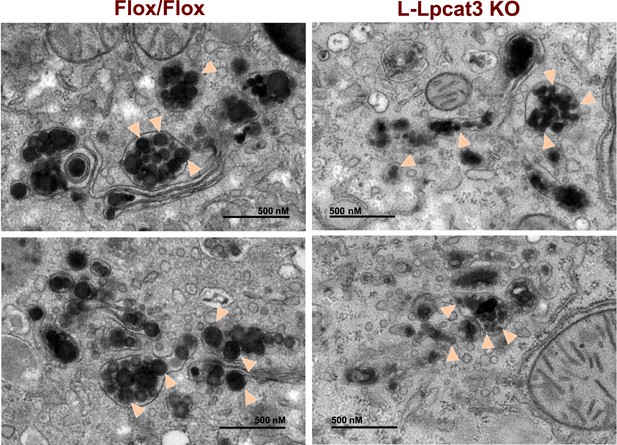
Reduced nascent lipoprotein particle size in the lumen of Golgi and secretory vesicles in Lpcat3-deficient liver.
EM of imidazole-stained liver sections. Lpcat3fl/fl (Fl/Fl) and Lpcat3fl/fl Albumin-Cre (L-Lpcat3 KO) mice were fasted for 6 hr. Samples were fixed and processed as described in the methods. Arrowheads indicate nascent lipoprotein particles.
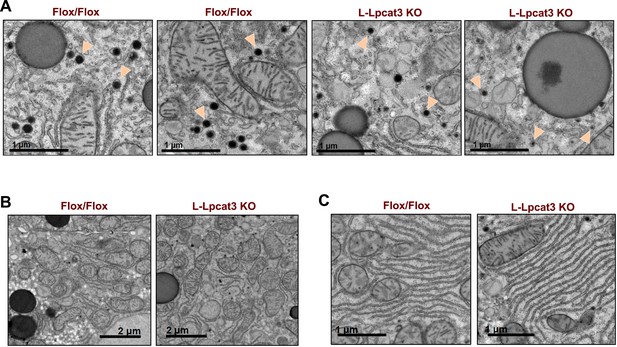
Loss of Lpcat3 in liver does not alter membrane structure in ER and mitochondria.
(A) EM of imidazole-stained liver sections. Arrowheads indicate nascent lipoprotein particles in smooth ER lumen. (B) EM of imidazole-stained liver sections. Mitochondria are shown. (C) EM of imidazole-stained liver sections. Rough ER is shown.
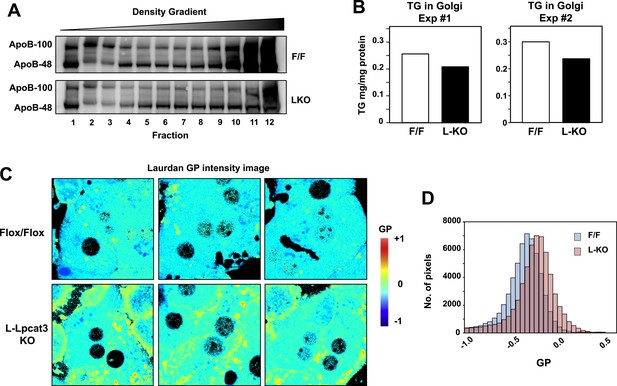
Lpcat3 activity regulates membrane lipid mobility and apoB lipidation in hepatocytes.
(A) The distribution of apoB-containing lipoproteins in the Golgi fractions isolated from livers of Lpcat3fl/fl (F/F) and Lpcat3fl/fl Albumin-Cre (LKO) mice. Golgi luminal contents were subject to a density gradient ultracentrifugation. 12 fractions from the gradient were harvested from top to bottom and analyzed by western blot. (B) Triglyceride contents of the Golgi fractions isolated from livers of Lpcat3fl/fl (F/F) and Lpcat3fl/fl Albumin-Cre (LKO) mice. Results from two representative experiments were shown. (C) Live primary hepatocytes from Lpcat3fl/fl (Flox/Flox) and Lpcat3fl/fl Albumin-Cre (L-Lpcat3 KO) mice were stained with laurdan. The laurdan emission spectrum was captured by a 2-photon laser-scan microscope. Generalized polarization (GP) was calculated from the emission intensities obtained from images. Higher GP value indicates that membranes are more ordered and less dynamic. The GP value of each pixel was used to generate a pseudocolor GP image. (D) The binary histograms of the GP distribution of the GP images (n = 4). The size of the GP binary is 0.05.
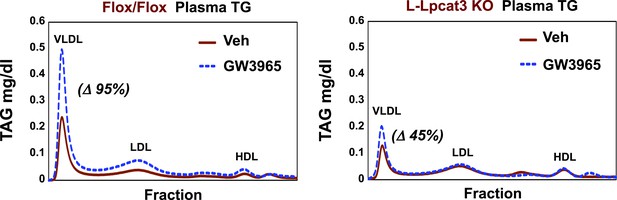
Lpcat3 is required for LXR-dependent VLDL TG secretion from liver.
Plasma lipoprotein profile from flox/flox and L-Lpcat3 mice gavaged with LXR agonist GW 3965 (20 mg/kg) for 2 days. Pooled plasma (5 mice/group) was analyzed by FPLC. Increases in VLDL TG were calculated based on the area under the curve.
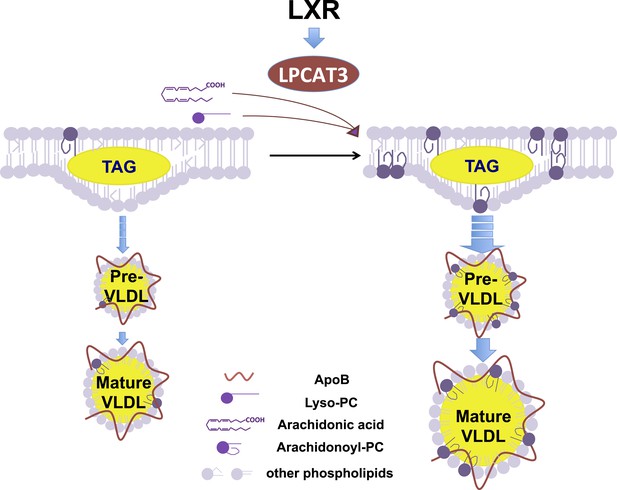
Schematic illustration of the role of the LXR-Lpcat3 pathway in VLDL lipidation.
Activation of LXRs promotes the incorporation of arachidonate into intracellular membranes through the induction of Lpcat3 expression. This change in membrane composition creates a dynamic membrane environment that facilitates the transfer of TG synthesized on ER and/or in cytosolic LD to nascent apoB–containing lipoproteins particles, leading to the efficient lipidation of apoB–containing lipoproteins.
Tables
Breeding data for global Lpcat3-deficient mice
| Genotype | Number of pups/mice | Observed % | Expected % | Time |
|---|---|---|---|---|
| WT | 25 | 28 | 25 | At birth |
| Het | 43 | 48 | 50 | |
| KO | 21 | 23 | 25 | |
| WT | 50 | 35 | 25 | At weaning |
| Het | 91 | 64 | 50 | |
| KO | 0 | 0 | 25 |
-
Genotypic ratio of newborns and weanlings obtained from Lpcat3 heterozygote intercrosses.
Breeding data for liver-specific Lpcat3-deficient mice
| Genotype | Number of mice | Observed % | Expected % |
|---|---|---|---|
| Flox/Flox | 26 | 53 | 50 |
| Flox/Floxalbumin-cre | 23 | 46 | 50 |
-
Genotypic ratio of adult mice obtained from Lpcat3fl/fl and Lpcat3fl/fl Albumin-Cre intercrosses.
Breeding data for intestine-specific Lpcat3-deficient mice
| Genotype | Number of pups/mice | Observed % | Expected % | Time |
|---|---|---|---|---|
| WT | 25 | 28 | 25 | At birth |
| Het | 43 | 48 | 50 | |
| KO | 21 | 23 | 25 |
| Genotype | Number of pups | Observed % | Expected % | Time |
|---|---|---|---|---|
| F/F, Cre+ | 20 | 24 | 25 | At birth |
| F/F, Cre− | 24 | 28 | 25 | |
| F/+, Cre+ | 19 | 22 | 25 | |
| F/+, Cre− | 20 | 24 | 25 | |
| F/F, Cre+ | 12 | 16 | 25 | 1 week old |
| F/F, Cre− | 24 | 32 | 25 | |
| F/+, Cre+ | 19 | 25 | 25 | |
| F/+, Cre− | 20 | 26 | 25 |
-
Genotypic ratio of newborns and 1 week-old pups obtained from Lpcat3fl/fl and Lpcat3+/F Villin-Cre intercrosses.
Quantitative real-time PCR primer sequences
| Murine qPCR primers | Forward | Reverse |
|---|---|---|
| CD36 | TTGAAAAGTCTCGGACATTGAG | TCAGATCCGAACACAGCGTA |
| PLPT | GTCTAAAATGAATATGGCCTTCG | CCAGAAGTGATGAACGTGGA |
| DGAT1 | TTCCGCCTCTGGGCATT | GCCCACAATCCAGGCCA |
| DGAT2 | GGCGCTACTTCCGAGACTAC | TGGTCAGCAGGTTGTGTGTC |
| APOA4 | ACCCAGCTAAGCAACAATGC | TGTCCTGGAAGAGGGTACTGA |
| APOE | GACTTGTTTCGGAAGGAGCTG | CCACTCGAGCTGATCTGTCA |
| AOPB | CTGAACATCAAGAGGGGCATC | GGTAACCTGAGTTGAGCAGTTT |
| MTTP | GGCAGTGCTTTTTCTCTGCT | TGAGAGGCCAGTTGTGTGAC |
| PDK4 | ACCGAAGAACCTGGCGAAG | TGATCCCGTAAAATGTCAGGC |
| CD68 | GACCTACATCAGAGCCCGAGT | CGCCATGAATGTCCACTG |
| CCL6 | TCTTTATCCTTGTGGCTGTCC | TGGAGGGTTATAGCGACGAT |
| ACSL3 | TCTAGGAGTGAAGGCCAACG | GCAATATCTGAGGGCAGTGG |
| ACSL5 | AACCAGTCTGTGGGGATTGAG | CGTCTTGGCGTCTGAGAAGTA |
| MOGAT2 | TCTTCCAGTACAGCTTTGGCCTCA | TGATATAGCGCTGATGAAGCCGGT |
| FABP1 | AGTACCAATTGCAGAGCCAGGAGA | GACAATGTCGCCCAATGTCATGGT |
| FABP2 | AGAGGAAGCTTGGAGCTCATGACA | TCGCTTGGCCTCAACTCCTTCATA |
| FATP1 | CGCTTTCTGCGTATCGTCTG | GATGCACGGGATCGTGTCT |
| LPCAT3 | GGCCTCTCAATTGCTTATTTCA | AGCACGACACATAGCAAGGA |
| LPCAT1 | GTGCACGAGCTGCGACT | GCTGCTCTGGCTCCTTATCA |
| LPCAT2 | TGTACTAATCGCTCCTGTTTGATT | CACTGGAACTCCTGGGATG |
| LPCAT4 | TTCGGTTTCAGAGGATACGACAA | AATGTCTGGATTGTCGGACTGAA |
| 36B4 | AGATGCAGCAGATCCGCAT | GTTCTTGCCCATCAGCACC |
| GAPDH | TGTGTCCGTCGTGGATCTGA | CCTGCTTCACCACCTTCTTGAT |





