Loss of the liver X receptor LXRα/β in peripheral sensory neurons modifies energy expenditure
Figures
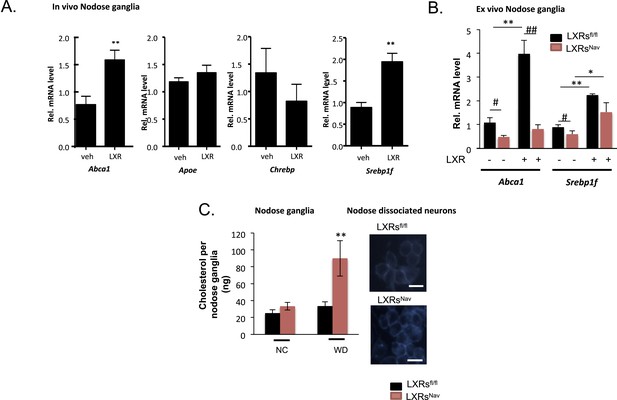
LXRs signaling regulates cholesterol metabolism in nodose ganglia neurons.
(A) Regulation of selected target genes in nodose ganglia (NG) following liver X receptor (LXR) agonist treatment in vivo (left panel) n = 5–8 per group. **p < 0.005. (B) NG organotypic slices were prepared from LXRsNav or LXRsfl/fl treated with GW3965 (5 μM) or vehicle for 4 hr. Quantitative PCR (qPCR) data are expressed as average fold-change relative to vehicle ± S.E.M., n = 3 independent experiments. # and *indicates p < 0.5, **p < 0.005. (C) Quantification of total cholesterol. Values were expressed as ng of cholesterol per NG, n = 6. ## and **p < 0.001. (right panel). Neurons isolated from LXRsNav or control mice NG were subjected to Filipin staining (representative images of staining from 3 individual mice).
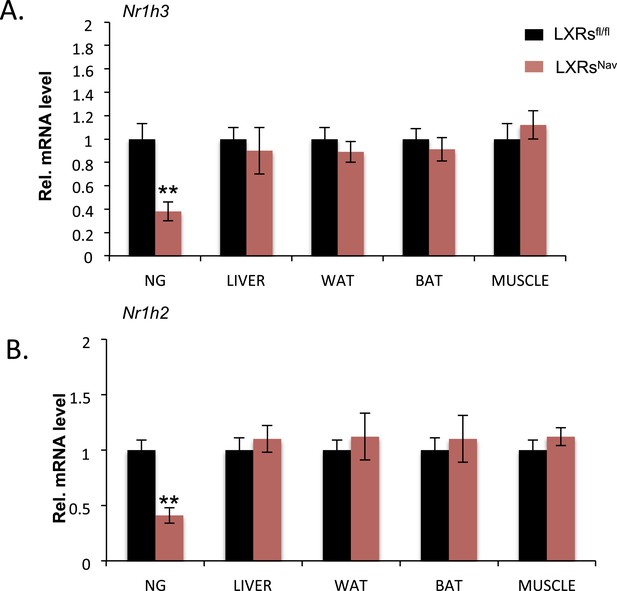
Ablation of LXR in the NAV1.8 positive neurons.
qPCR analysis detecting the expression of truncated isoforms of Nr1h3 (A) and Nr1h2 (B) in NG, liver, white adipose, brown adipose tissues (BATs), and muscle. Error bars show S.E.M. **indicates p < 0.01.
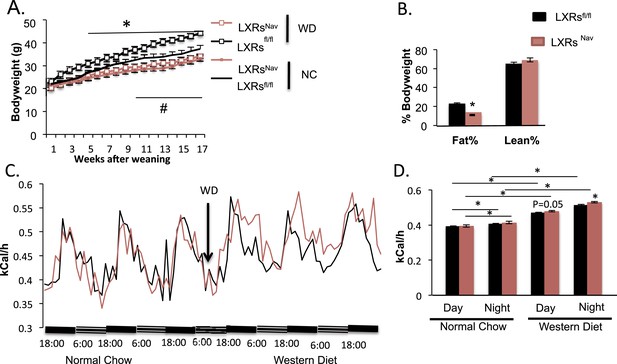
Ablation of LXRs from NAV1.8 expressing neurons exacerbates high-fat diet induced thermogenesis.
(A) Body weight of LXRsNav mice and littermate controls on chow and Western Diet (WD) as followed over time (n = 12 per genotype) * for WD, # for normal chow (NC). (B) Adipose tissue as a percentage of total body weight in mice fed WD for 9 weeks, measured by NMR (n = 6). (C,D) Energy expenditure in weight-matched mice. (C) Calorimetry trace before and after a switch from NC to WD. (D) Energy expenditure during light and dark cycles before and after a switch from NC to WD. Error bars show S.E.M. *indicates p < 0.05.
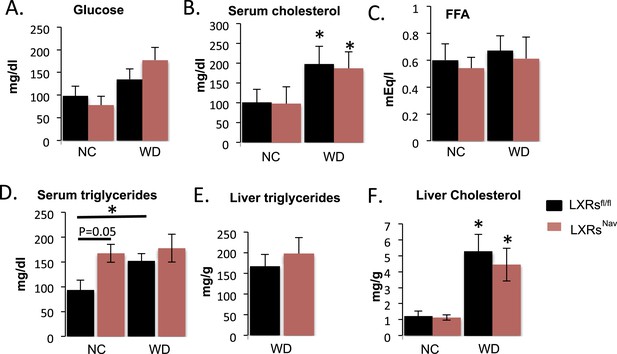
Blood chemistry and lipid levels of LXRsNav mice.
Metabolite levels as measured by the UTSW metabolic core from LXRsNav and LXRsfl/fl mice fed 16 weeks with NC or WD. (A) Glucose. (B) Serum cholesterol. (C) Fatty acid (FFA). (D) Serum triglycerides. (E) Liver triglycerides. (F) Liver cholesterol.
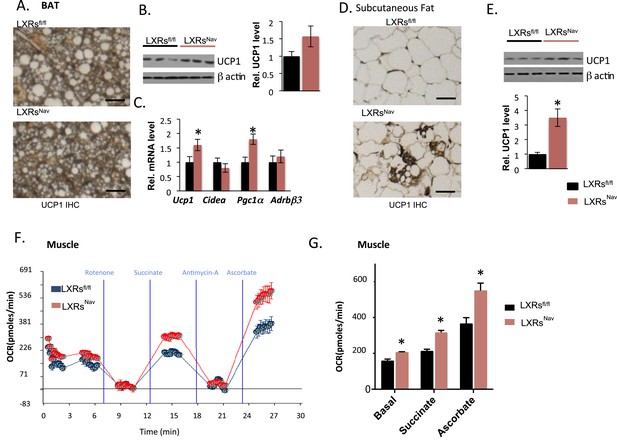
Adipose tissue and muscle reprogramming in LXRsNav mice vs control mice.
(A) Immunohistochemistry for UCP1 in BAT. (B) UCP1 Western blot on whole BAT pads (upper panel, n = 4) UCP1 molecular weight (MW) is 33 kDa, Actin served as the loading control, MW 42 kDa, the graph represents blot quantification. (C) mRNA levels in the BAT of LXR ablated mice vs control mice (lower panel, n = 4). *indicates p < 0.05. (D) UCP1 staining and (E) Western-blot analysis of UCP1 protein levels in whole, individual dorsal subcutaneous fat pads. Actin served as the loading control. The signal is quantified in the graph below (n = 4) Scale bar = 100 μm. (F,G) Oxygen-consumption rates were determined using the XF24 Extracellular Flux Analyzer following the manufacturers' protocols (n = 3). *indicates p < 0.01.
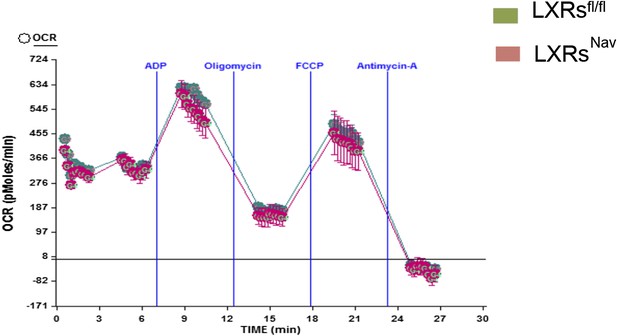
Electron-flow experiments.
Isolated skeletal muscle mitochondria were seeded at 10 μg of protein per well in XF24 V7 cell-culture microplates and analyzed with the XF24 Extracellular Flux Analyzer (n = 3).
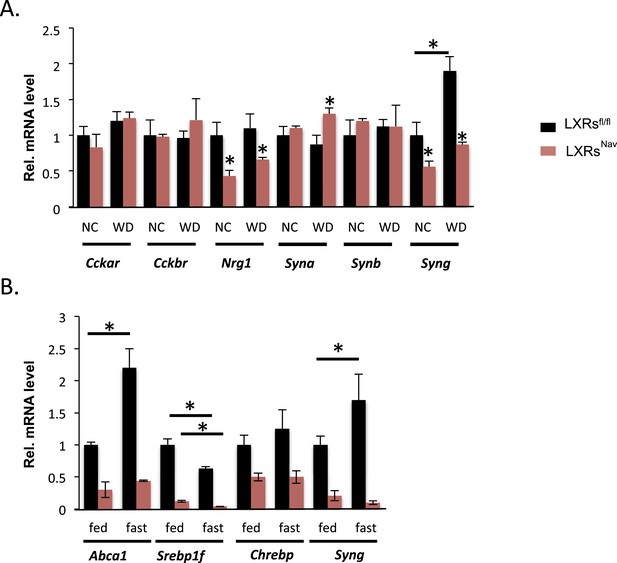
NG gene expression in LXRsNav mice vs control mice.
(A) Gene expression in NG from LXRsNav and control mice fed with NC or WD. (B) Gene expression in NG from LXRsNav and control mice fasted 20 hr or fed. All genes show a significant difference between both genotypes. *indicates p < 0.05 (n = 6–10).





