Experimental evolution reveals hidden diversity in evolutionary pathways
Figures
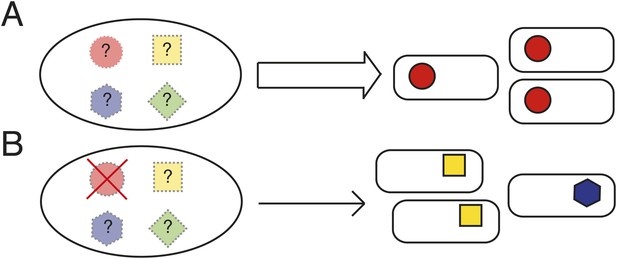
Determining the causes of parallel evolution.
(A) A selective challenge repeatedly leads to a phenotypic adaptation with the same mutational solution represented by red circles. If only one solution is ever observed (the red circles), how can it be proven that alternative pathways exist (the unrealized [shaded] mutational pathways marked with ‘?’)? (B) In an experimental system where common genetic pathways to an adaptive phenotype can be removed (the red circle with a cross), alternative pathways (blue and yellow) can be revealed and the underlying causes of the parallel evolution determined.
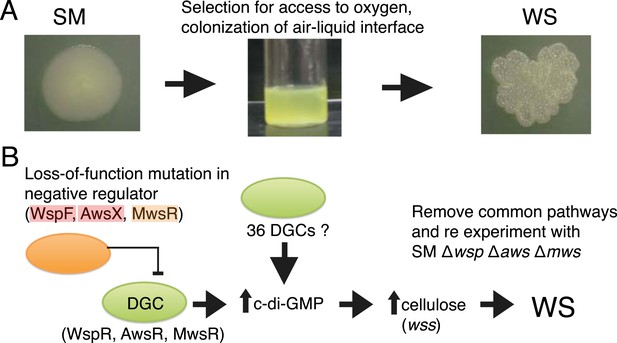
The Pseudomonas fluorescens SBW25 wrinkly spreader model.
(A) The ancestral smooth (SM) strain evolves to colonize the air–liquid interface of a static microcosm. The ability to make a mat at the air–liquid interface is dependent on mutational activation of the gene products of the wss operon, which encodes the biosynthetic machinery for production of cellulose that function as an extracellular glue (Spiers et al., 2003). These mat-forming types display a wrinkled morphology on agar plates and are referred to as wrinkly spreaders (WS). Over-activation of cellulose production is caused by an increase in the second messenger c-di-GMP—the product of diguanylate cyclases (DGCs) (Goymer et al., 2006; Malone et al., 2007). (B) All WS mutants described in previous work have mutations in one of three loci (wsp, aws, and mws), all involving DGCs under negative regulation (McDonald et al., 2009). There are 36 additional DGCs in the genome, and the ability to remove the three commonly followed pathways makes it possible to determine whether evolution can follow alternate routes to the WS phenotype.
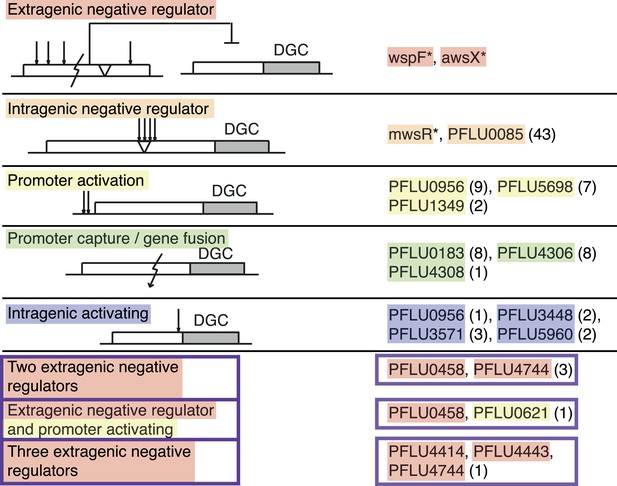
Mutational routes to WS.
Numbers in parentheses are the number of independent mutants found out of 91. Putative functional effects of mutations are based on mutational patterns. Classification of mutational types has been colour coded: Red, extragenic negative regulators; orange, intragenic negative regulators; yellow, promoters (activating mutations); green, gene fusion/promoter capture; blue, intragenic (activating mutations); and purple, double and triple mutations.
-
Figure 3—source data 1
Full genetic data for all new WS mutants.
- https://doi.org/10.7554/eLife.07074.006
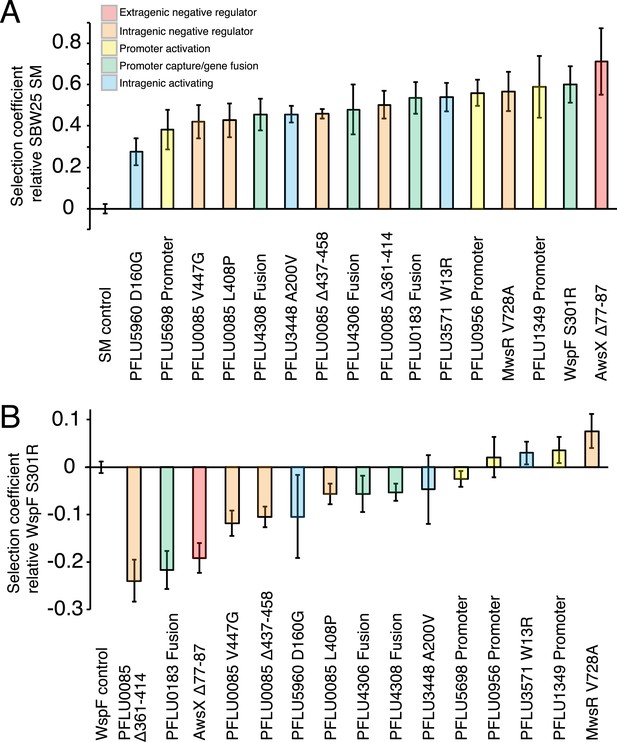
Fitness effects of WS types arising from single mutations.
(A) Fitness in a 1:100 invasion assay against the ancestral SM genotypes. (B) Fitness in a 1:1 competitive assay against the high fitness LSWS (WspF S301R) genotype. Error bars represent SD (n = 8).
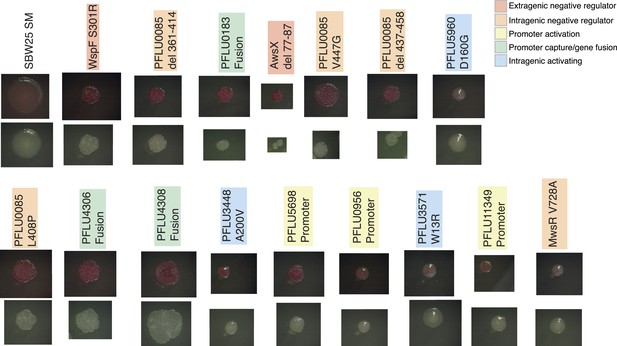
Colony morphology of WS types arising from single mutations on KB agar with and without Congo Red.
https://doi.org/10.7554/eLife.07074.009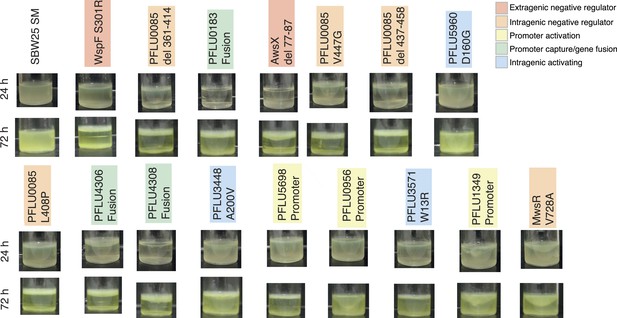
Microcosms of WS types arising from single mutations after 24 hr and 72 hr static growth.
https://doi.org/10.7554/eLife.07074.010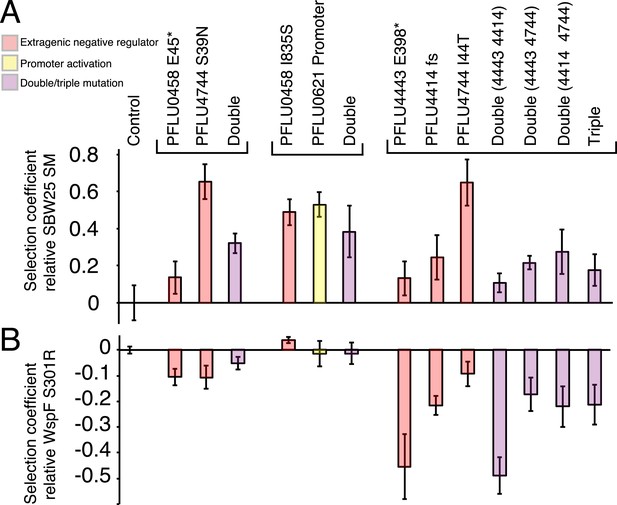
Fitness effects of WS types arising from two and three mutational events.
Individual mutations were recreated in the ancestral SM genotype plus combinations of mutations (see text). (A) Fitness in a 1:100 invasion assay against the ancestral SM genotypes. (B) Fitness in a 1:1 competitive assay against the high fitness LSWS (WspF S301R) genotype. Error bars represent SD (n = 8).
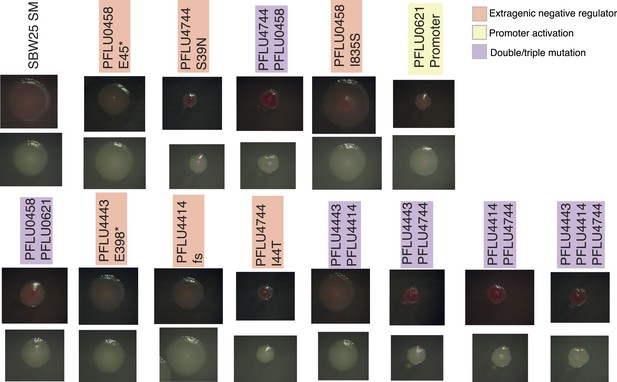
Colony morphology of WS types arising from two and three mutational events on KB agar with and without Congo Red.
https://doi.org/10.7554/eLife.07074.012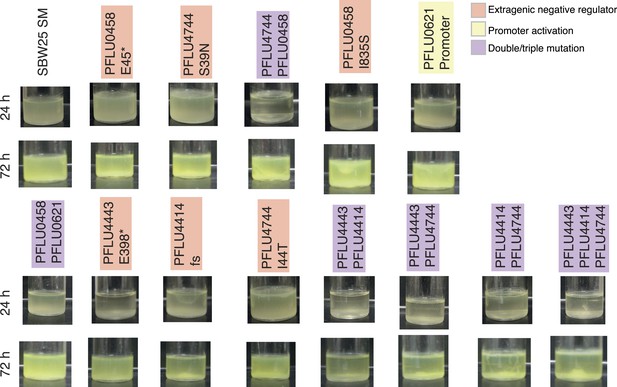
Microcosms of WS types arising from two and three mutational events after 24 hr and 72 hr static growth.
https://doi.org/10.7554/eLife.07074.013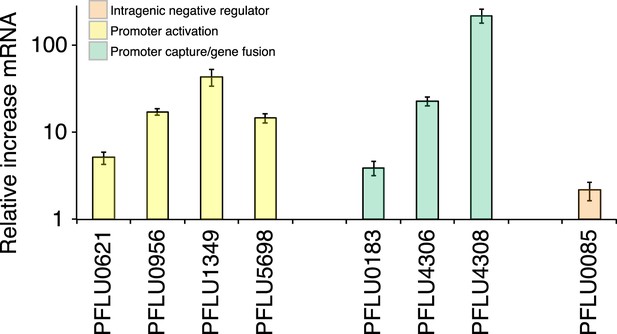
Relative increase of mRNA levels in WS mutants with putative promoter or gene fusion mutations compared to SM SBW25 measured by quantitative PCR.
PFLU0085 is the Δ361–414 deletion mutant not expected to result in increased mRNA levels used as a control. Error bars represent SEM.
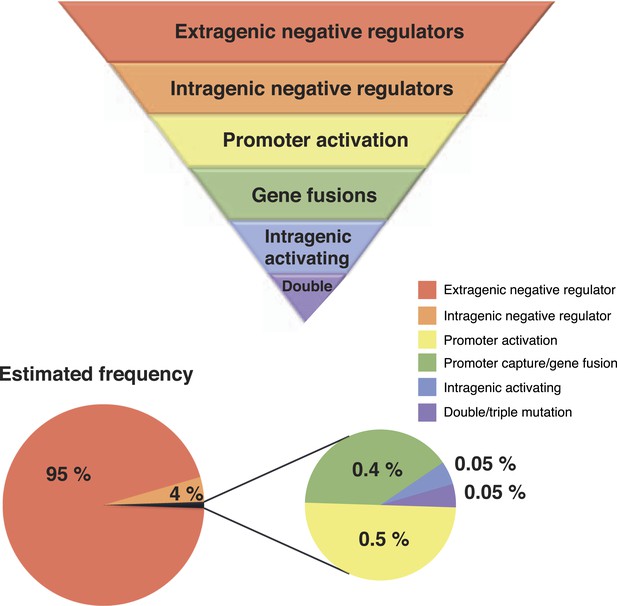
Hierarchical principles of mutational gene activation.
Certain pathways have a greater capacity than others to translate mutation into phenotypic variation depending on the functional and regulatory interactions involved. The estimated number of mutants is a first approximation of how mutations are expected to be distributed between different categories assuming a similar number of target genes in each category. This distribution can also be biased by mutational hot spots and differences in fitness effects of mutations in the gene involved.
Tables
Summary of mutations in different categories
| Category | Gene locus | Types of mutation | Number of mutants |
|---|---|---|---|
| Extragenic negative regulator | wspF (PFLU1224) | deletion, insertion, stop, frameshift, amino acid substitution | previous work* |
| awsX (PFLU5211) | deletion, insertion, stop, frameshift, amino acid substitution | previous work* | |
| Intragenic negative regulator | mwsR (PFLU5329) | deletion, insertion, amino acid substitution | previous work* |
| PFLU0085 | deletion, insertion, amino acid substitution | 43 | |
| Promoter activating | PFLU0956 | substitutions, small deletion and insertions | 9 |
| PFLU5698 | substitutions, small insertion | 7 | |
| PFLU1349 | deletion, small insertion | 2 | |
| Promoter capture/gene fusion | PFLU0183 | deletion | 8 |
| PFLU4306 | deletion | 8 | |
| PFLU4308 | deletion | 1 | |
| Intragenic activating | PFLU0956 | amino acid substitution | 1 |
| PFLU3448 | amino acid substitution | 2 | |
| PFLU3571 | amino acid substitution | 3 | |
| PFLU5960 | amino acid substitution | 2 | |
| Double and triple mutants | PFLU0458 | amino acid substitution, stop codon, frameshift | 3 |
| PFLU4744 | amino acid substitution, frame shift | ||
| PFLU0458 | amino acid substitution | 1 | |
| PFLU0621 | substitution (promoter) | ||
| PFLU4414 | frame shift | 1 | |
| PFLU4443 | stop | ||
| PFLU4744 | amino acid substitution |
-
*
Mutational targets described in McDonald et al. (2009).
Additional files
-
Supplementary file 1
Phyre2 pdb model of PFLU5960.
- https://doi.org/10.7554/eLife.07074.016
-
Supplementary file 2
Oligonucleotide primers.
- https://doi.org/10.7554/eLife.07074.017





