Structural basis for recognition and remodeling of the TBP:DNA:NC2 complex by Mot1
Figures
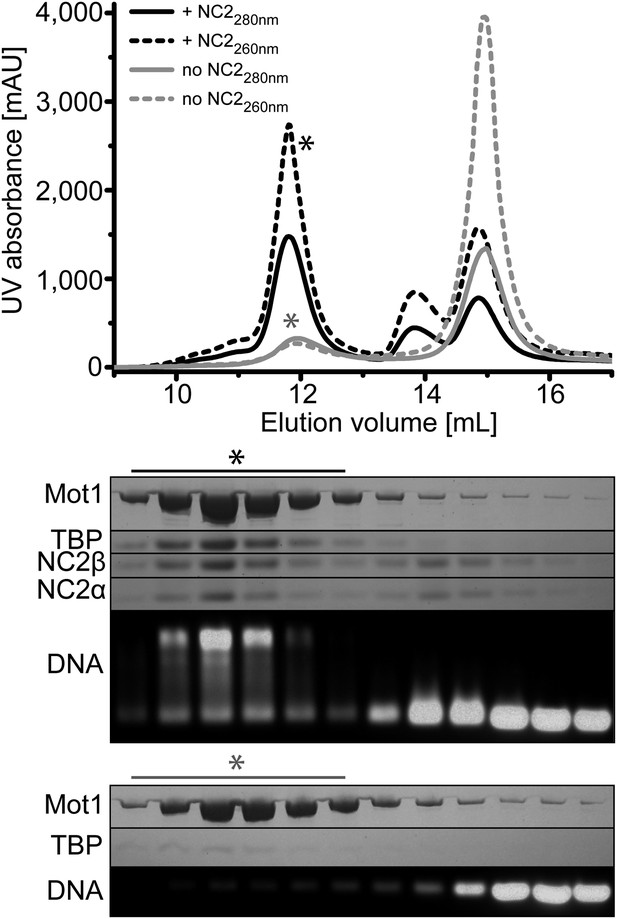
Size exclusion chromatography of the E. cuniculi Mot1:TBP:DNA:NC2 complex.
Upper panel: elution profiles of Mot1:TBP:DNA complex (gray) and Mot1:TBP:DNA:NC2 complex (black). Absorbance at 260 nm is represented by dashed lines and at 280 nm as solid lines. Lower panel: analysis by SDS-PAGE (Coomassie staining) and agarose gel electrophoresis (Gel-Red staining). The asterisk marks fractions containing all components.
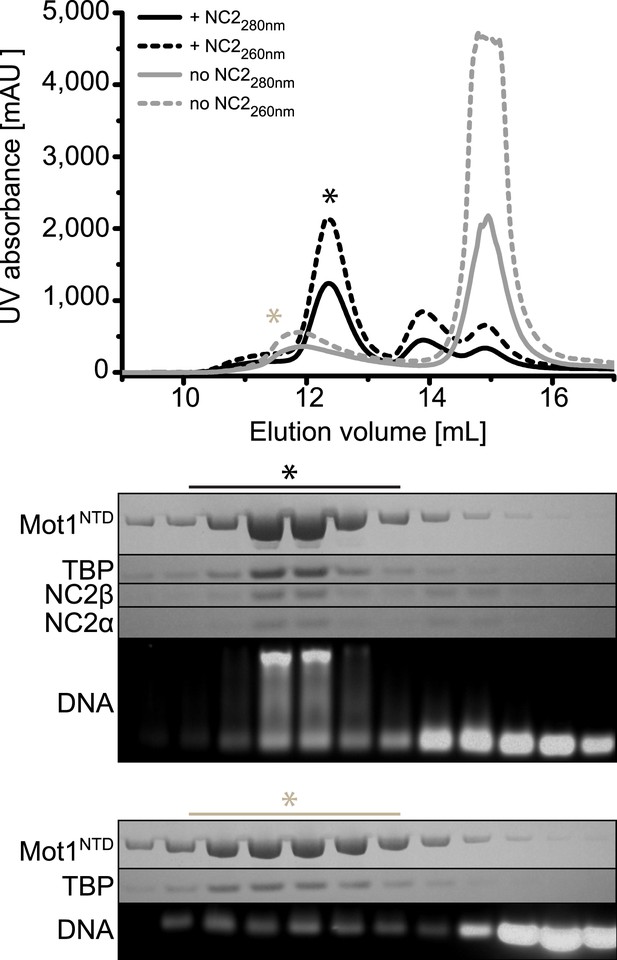
Size exclusion chromatography of the E. cuniculi Mot1NTD:TBP:DNA:NC2 complex.
Upper panel: elution profiles of Mot1NTD:TBP:DNA complex (gray) and Mot1NTD:TBP:DNA:NC2 complex (black) in size exclusion chromatography. Absorbance at 260 nm is represented as dashed lines and at 280 nm as solid lines. Lower panel: analysis of the co-purified complexes by SDS-PAGE (Coomassie) and agarose gel electrophoresis (Gel-Red).
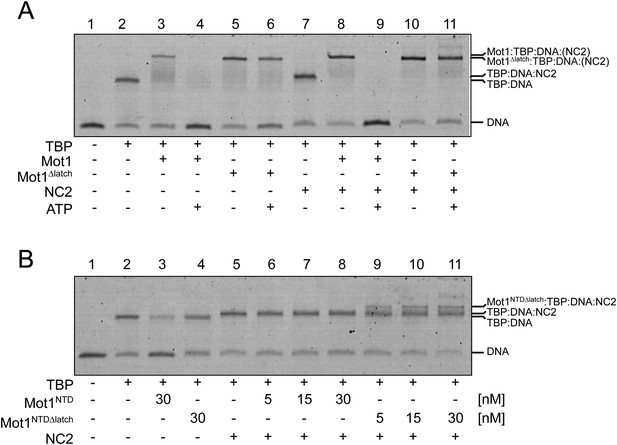
Electrophoretic mobility shift analysis of Mot1:TBP:DNA:NC2 complexes.
(A) Upon ATP addition, Mot1 dissociated TBP from DNA (lanes 3 and 4) as well as the TBP:DNA:NC2 complex (lanes 8 and 9). Mot1Δlatch was impaired in TBP removal (lanes 5 vs 6 and 10 vs 11). (B) Effect of NC2 on ATP-independent remodeling by Mot1NTD. Addition of NC2 prevented the Mot1NTD from displacing TBP from DNA (lane 3 and 6–8). The Mot1NTDΔlatch protein bound TBP:DNA more stably than Mot1NTD (lane 4). Addition of NC2 to Mot1NTDΔlatch:TBP:DNA complex resulted in a distinct shift (lanes 9–11), consistent with formation of a stable complex containing all components.
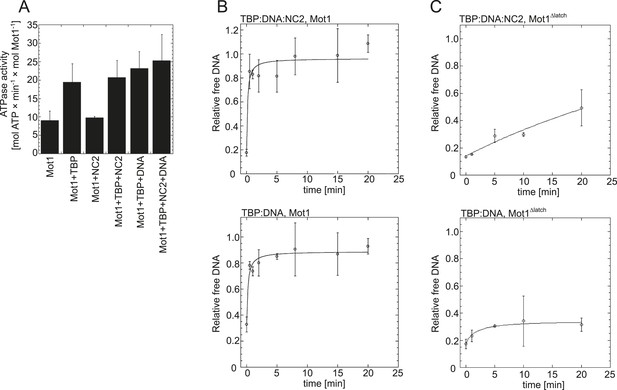
Effect of NC2, TBP, and DNA on Mot1's activity.
(A) The graph shows the steady-state ATPase activity of 8 nM Mot1 alone or in the presence of 80 nM NC2 and/or TBP and with or without 23 nM TATA-containing DNA. The assays were performed as described previously (Wollmann et al., 2011). The data represent the mean ± the standard deviation obtained from at least two independent experiments. (B) The graphs show the rates of Mot1-catalyzed TBP:DNA:NC2 dissociation (top) and TBP:DNA dissociation (bottom) measured by quantitation of the free DNA level by EMSA following ATP addition to pre-formed complexes. The assays were performed as previously described (Wollmann et al., 2011). (C) The same as in (B) shown for the Mot1Δlatch mutant.
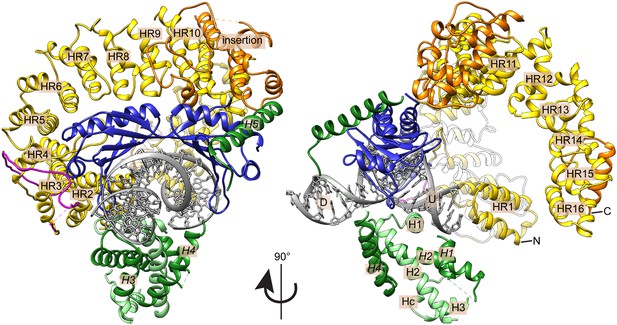
Crystal structure of the E. cuniculi Mot1NTD:TBP:DNA:NC2 complex.
Front and side views of the structure, represented as cartoon models with highlighted secondary structure. TBP (blue), NC2α (light green), and NC2β (dark green) encircle kinked promoter DNA (gray with upstream (U) and downstream (D) regions labeled). The HEAT repeats (HR, yellow with N- and C-termini marked) and insertion domain (orange) of the Mot1NTD bind along the convex surface of TBP and contact the C-terminal helix (H5) of NC2β. The latch of Mot1NTD (magenta), which has been previously shown to bind to TBP's DNA-binding cleft, is mostly disordered in the presence of DNA and NC2.
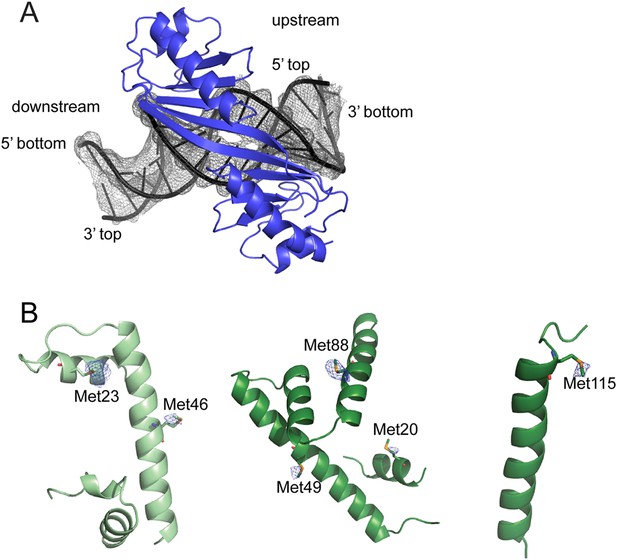
X-ray electron density maps.
(A) 2Fo – Fc electron density map of DNA bases displayed at 1σ contour level (gray mesh). TBP is shown in blue. The widening at the TATA box is nicely visible. (B) Anomalous density map (blue mesh) contoured at 3σ level shows signal of some of the selenium sites of NC2α (left), NC2β residues 12–101 (middle) and NC2β residues 110–137 (right).
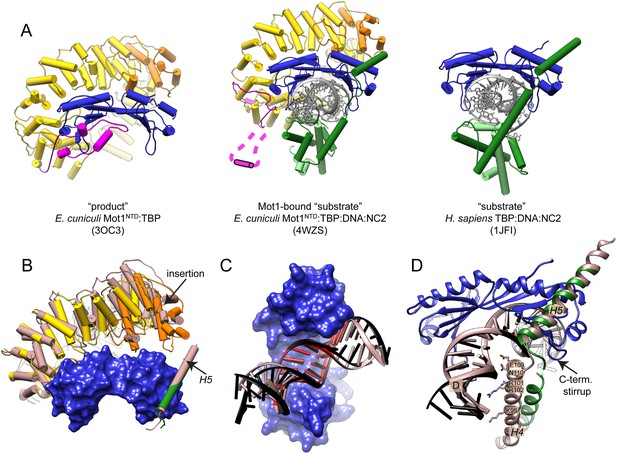
Features of the Mot1-bound ‘substrate’ complex.
(A) Comparison of the ‘product’ (left, Wollmann et al., 2011), ‘substrate’ (right, Kamada et al., 2001), and Mot1-bound ‛substrate’ complex (center). Mot1's latch in the latter structure is disordered and represented schematically. Color code is as in Figure 4. Panel (B) shows Mot1NTD and TBP from the Mot1NTD:TBP:DNA:NC2 structure (color code as in Figure 4) superimposed via TBP with the ‘product’ Mot1NTD:TBP structure and NC2β H5 from the ‘substrate’ TBP:DNA:NC2 structure (both shown in light brown). Mot1NTD shifts toward H5 and TBP although the position of H5 is not affected. Panels (C) and (D) show conformational changes of NC2 and DNA in the Mot1NTD:TBP:DNA:NC2 complex structure (color code as in Figure 4) compared to the TBP:DNA:NC2 ‘substrate’ complex (shown in light brown) superimposed via TBP. (C) View from the concave side of TBP shows that in the presence of Mot1, DNA is partially straightened and underwound. TATA box region is highlighted in red. Panel (D) shows that partially unfolded helix H4 of NC2β, which joins the NC2HF with helix H5, loses its interaction with downstream DNA and is close to TBP's C-terminal stirrup.
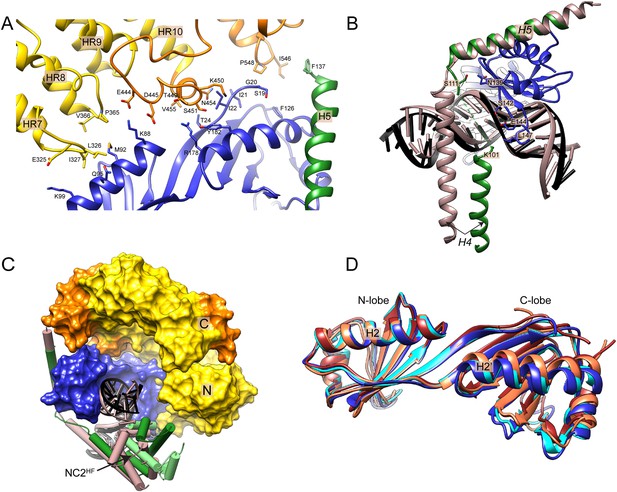
Mot1NTD:TBP:DNA:NC2 structure features.
Panel (A) shows the interface between Mot1NTD HR 7–10, TBP and H5 of NC2β. Key residues are annotated. Panel (B) shows helix H4 of NC2β, which is in a direct proximity to TBP’s C-terminal stirrup. Part of the chain is unfolded. Color code of Mot1NTD:TBP:DNA:NC2 is as in Figure 5, ‘substrate’ TBP:DNA:NC2 complex is shown in light brown. (C) View along the upstream DNA shows that Mot1NTD binding induces NC2HF and upstream DNA to shift towards the N- and C-terminus of Mot1NTD (N and C). (D) Comparison of TBP’s conformation in the E. cuniculi TBP dimer (coral), E. cuniculi Mot1NTD:TBP complex (cyan), H. sapiens TBP:DNA:NC2 complex (brown) and the E. cuniculi Mot1NTD:TBP:DNA:NC2 complex (dark blue) crystal structures superposed via TBP’s helix H2.
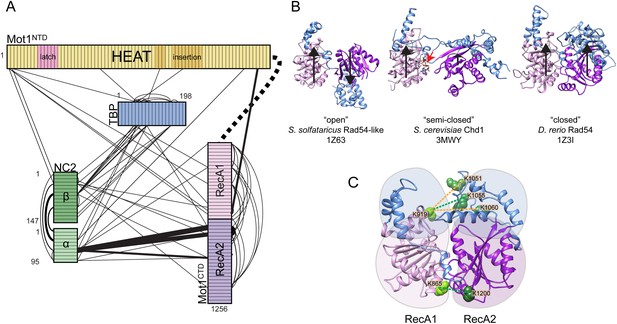
Mot1:TBP:DNA:NC2 complex analyzed by CX-MS.
(A) General topology of the Mot1:TBP:DNA:NC2 complex in the presence of ATPγS derived from the CX-MS data. Each polypeptide is divided into 10 amino acid segments. The black solid lines represent the identified cross-links. For simplicity, cross-links to the latch region of Mot1 are not displayed. Line thickness is proportional to the number of cross-links detected between joined segments. The dashed line between Mot1NTD and Mot1CTD represents the eight amino acid linker between these domains. (B) Crystal structures of Swi2/Snf2 domains used for the cross-link analysis: Sulfolobus solfataricus Rad54-like in an ‘open’ conformation (Dürr et al., 2005), Saccharomyces cerevisiae Chd1 in a ‘semi-closed’ conformation (Hauk et al., 2010) in the presence of ATPγS (red arrow) and Danio rerio Rad54 in a ‘closed’ conformation (Thomä et al., 2005). PDB accession codes are included. The structures are oriented with respect to the RecA1 subdomain. Relative orientations of the RecA1 (pink) and RecA2 (purple) subdomains are represented by the black arrows. Family-specific insertion regions are indicated in blue. Auxiliary domains were omitted. Panel (C) shows cross-linked sites listed in Table 3 part C mapped on the Swi2/Snf2 domain modeled in the ‘closed’ conformation. Cross-links detected only in the presence of ADP·BeFx and ATPγS are shown as green dashed lines, whereas the cross-links present only in the ADP·BeFx data set are in orange.
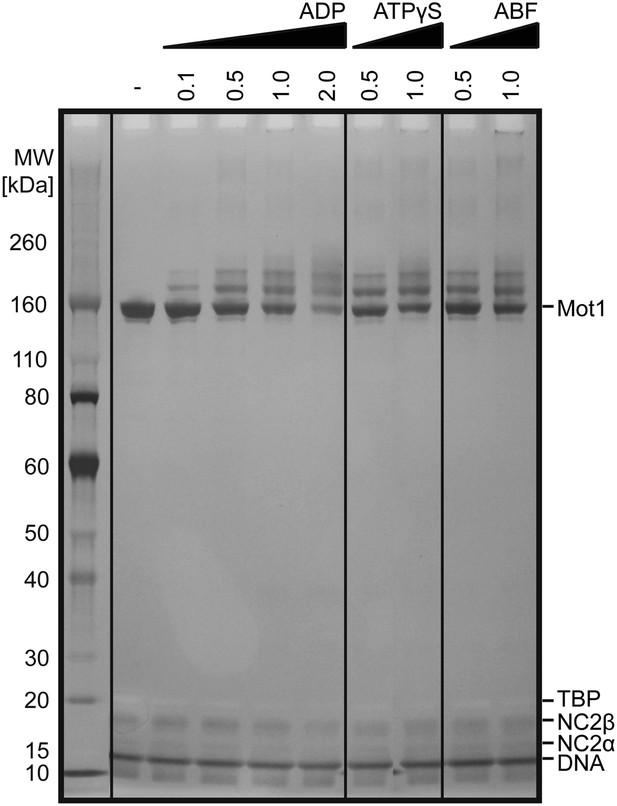
Titration of the cross-linking agent disulfosuccinimidyl glutarate (DSSG).
Cross-linking was performed on the Mot1:TBP:DNA:NC2 complex in the presence of ADP, ATPγS or ADP·BeFx (ABF). Numbers correspond to the molar excess of the cross-linking agent over lysine residues. A molar ratio of 1.0 was used to obtain the cross-linking data reported here.
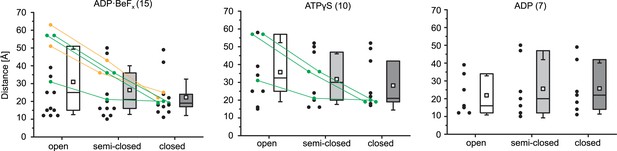
Analysis of the cross-links between RecA1 and RecA2 subdomains within the Mot1CTD models.
Numbers in brackets refer to the total number of obtained inter-subdomain cross-links. Data points marked in green represent the cross-links detected only in the ATPγS and ADP·BeFx data sets, whereas the data points shown in orange represent the cross-links detected exclusively in the ADP·BeFx data set. The colored rectangles include distances within the first to the third quartile with the median value indicated as line. The whiskers represent one standard deviation above and below the mean (open square).
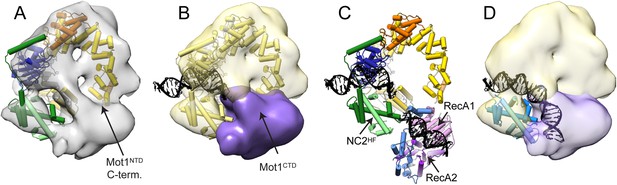
Pseudoatomic model for the Mot1:TBP:DNA:NC2 complex.
Panels (A) and (B) show the Mot1NTD:TBP:DNA:NC2 crystal structure rigid body docked into the negative stain EM map of the Mot1:TBP:DNA:NC2:ADP·BeFx complex. (B) The density segment corresponding to Mot1CTD (purple) is in a direct proximity to the C-terminal end of Mot1NTD and to NC2HF. The transparent yellow segment corresponds to the Mot1NTD:TBP:NC2 module. The TBP-bound promoter DNA fragment from the crystal structure is included (black). (C) Orientation of Mot1CTD derived from the interpretation of the CX-MS data. For simplicity, one of the best-scoring models is shown. RecA1 (pink) and RecA2 (purple) correspond to the subdomains of a Swi2/Snf2 fold. Protrusions are indicated in blue. The DNA fragment bound to Mot1CTD is modeled based on the SsoRad54-like:DNA crystal structure by superimposing via the RecA1 subdomain (Dürr et al., 2005). (D) Overlay of the EM map segments from (B) and the crystal structure of NF-YB/NF-YC transcription factor (blue) bound to a DNA fragment (Nardini et al., 2013) superimposed via the histone fold of NC2. Color coding for Mot1NTD, TBP, and NC2 shown in (A) and (C) is the same as in Figure 4.
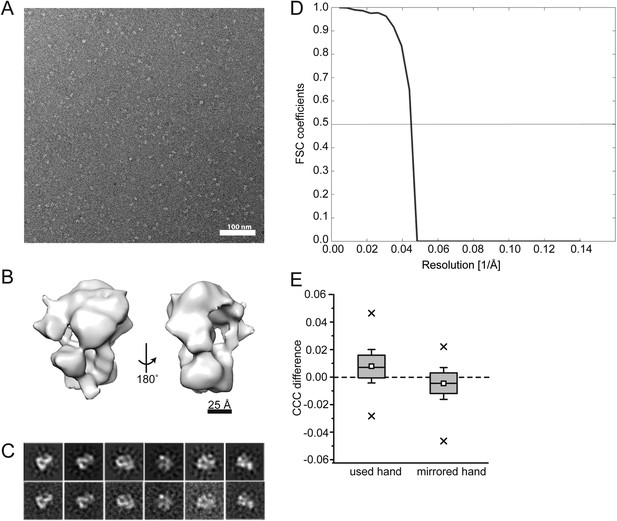
EM data.
(A) Negative stain micrograph of the E. cuniculi Mot1:TBP:DNA:NC2:ADP·BeFx complex. (B) Electron density map of the 22 Å negative stain reconstruction. The map was contoured at ∼300,000 Å3, which corresponds to the theoretical volume of the components calculated for the atomic models. (C) 2D class averages (bottom) and the back projections of the 3D model in the Euler angle directions assigned to the class averages (top). (D) Fourier shell correlation curve of the calculated reconstruction filtered at 20 Å. (E) Experimental determination of the absolute hand of the Mot1:TBP:DNA:NC2:ADP:BeFx complex. The difference of the cross correlation coefficient values of the template matching on a negative stain tomogram with both hands is plotted (n = 124). Positive values for the used hand indicate a higher agreement of the determined map with the particles in the tomogram than with its mirrored version. Consequently, the mirrored hand has a negative CCC difference compared with the real hand. Thus, the hand of the discussed and interpreted map is correct. Additionally, this test is a validation of the map itself, as only a correct map could lead to a discrimination of the handedness. Box plot description is same as in Figure 6 (p < 0.05 in two-sample t–test).
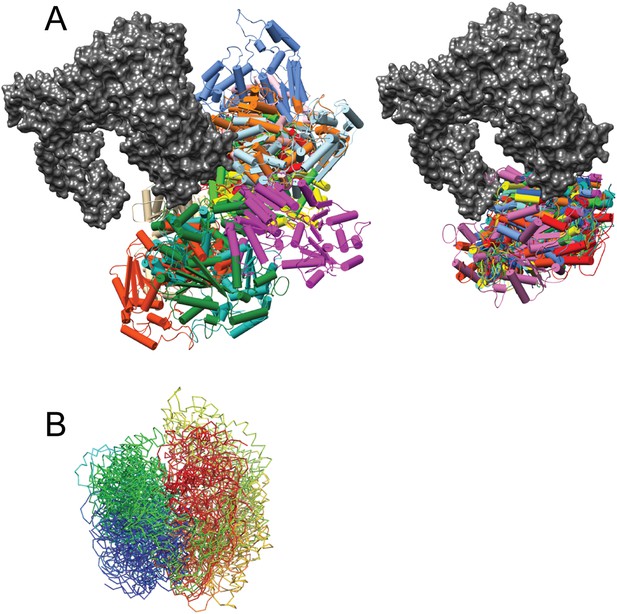
Analysis of Mot1CTD localization based on the CX-MS data.
(A) Overlay of 12 randomly selected (left) and 12 best-scoring Mot1CTD positions (right) presented in different colors superimposed via Mot1NTD:TBP:DNA:NC2 crystal structure (gray surface). (B) Overlay of the best-scoring Mot1CTD models shown in (A) from N- to C-terminus (rainbow-colored backbone representation) demonstrates that not only the locations but also the orientations of the best-scoring models are closely related to each other.
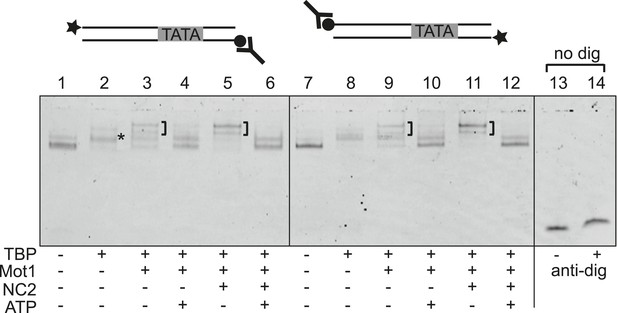
Mot1-mediated displacement of TBP and NC2 from end-blocked DNA templates.
Electrophoretic mobility shift analysis of ATP-dependent disruption of Mot1:TBP:DNA complexes with and without NC2 as in Figure 2, but using DNA substrates that carry a digoxygenin label at one end (circle) and fluorescein for DNA detection at the other end (star). Reactions in lanes 13 and 14 were performed using DNA alone without digoxygenin modification, and demonstrate that the addition of digoxygenin antibody (Y-shape) had no effect on the mobility of unmodified DNA. Blocking of either end by the antibody did not result in a detectable decrease in TBP dissociation activity, suggesting that Mot1 does not translocate an intact TBP:NC2 clamp along DNA but locally disrupts the TBP:DNA:NC2 complex.
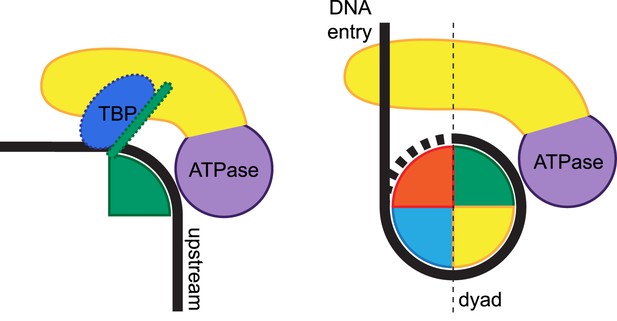
Remodeling of substrate protein:DNA complexes by Mot1 and comparison to the ISWI-type nucleosome remodeler.
Left side: the binding of Mot1NTD (yellow) induces destabilization of TBP:DNA and NC2:DNA interactions (TBP is shown in blue, NC2 is represented by a green shape). Right side: an ISWI-type Swi2/Snf2 remodeler bound to nucleosome according to current models (Dang and Bartholomew, 2007; Yamada et al., 2011; Hota et al., 2013). The DNA-binding domain (yellow) engages extranucleosomal DNA at the entry site. In both cases, the Swi2/Snf2 ATPase domains (purple) specifically recognize their histone fold:DNA substrates.
Tables
Data collection and refinement statistics
| Data collection* | |
| Space group | C 1 2 1 |
| Cell dimensions | |
| a, b, c (Å) | 150.6, 140.3, 90.8 |
| α, β, γ (°) | 90.0, 113.7, 90.0 |
| Resolution (Å) | 49.2–3.8 (4.0–3.8)† |
| Rmerge (%) | 10.4 (78.9) |
| CC(1/2) | 99.8 (83.8) |
| I/σI | 7.5 (1.5) |
| Completeness (%) | 98.2 (93.5) |
| Redundancy | 3.4 (3.4) |
| Refinement | |
| Resolution (Å) | 49.2–3.0 (4.0–3.8) |
| No. reflections | 17,163 |
| Rwork/Rfree (%) | 23.5/25.8 (26.9/30.2) |
| No. atoms | |
| Protein | 8422 |
| DNA | 799 |
| Ligand/ion | 0 |
| Water | 0 |
| Isotropic B-factors (Å2) | |
| Protein | 69 |
| DNA | 135 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 0.75 |
-
*
From one crystal.
-
†
Values in parentheses are for the highest-resolution shell.
Localization of the cross-links identified in Mot1:TBP:DNA:NC2 complex
| Experiment | Crystal structure | Within Mot1CTD | Latch-crystal structure | Latch– Mot1CTD | Crystal structure– Mot1CTD | Total | Decoy§ | Estimated FDR [%] | |
|---|---|---|---|---|---|---|---|---|---|
| Intralobe* | Interlobe† | ||||||||
| ADP·BeFx | 46 (42) | 21 (17) | 15 (12) | 8 (8) | 5 (5) | 37 (31) + 1 (1)‡ | 133 (116) | 2 | 0.8 |
| ATPγS | 51 (44) | 17 (14) | 10 (9) | 11 (11) | 3 (3) | 37 (28) | 129 (109) | 1 | 1.5 |
| ADP | 40 (36) | 14 (11) | 7 (7) | 11 (10) | 4 (3) | 21 (15) | 97 (82) | 2 | 2.0 |
-
*
Within RecA1 or RecA2 subdomain.
-
†
Between RecA1 and RecA2 subdomain.
-
‡
Between Mot1CTD and the linker joining N- and C-terminal domains (could not be mapped).
-
§
Detected from a reverse database, estimating false-discovery rate.
-
Numbers refer to the total number of cross links, including cross-linked sites which were detected more than once (i.e., from miss-cleaved peptides). Numbers in brackets refer to non-redundant linkages only.
-
Table 2—source data 1
Full list of the detected cross-links.
- https://doi.org/10.7554/eLife.07432.013
Distances of the cross-links detected between RecA1 and RecA2 subdomains of Mot1CTD mapped on different structural models
| Detected linkages | Euclidean Cα–Cα distance [Å] | Total number of detected cross links | ||||||
|---|---|---|---|---|---|---|---|---|
| Residue 1 | Residue 2 | SsoRad54-like (‘open’) | DrRad54 (‘closed’) | ScChd1 (‘semi-closed’) | ADP·BeFx | ATPγS | ADP | |
| A | 796 | 1013 | 16 | 22 | 20 | 1 | 1 | 1 |
| 796 | 1200 | 25 | 17 | 16 | 1 | 1 | 0 | |
| 842 | 1055 | 58 | 52 | 52 | 0 | 1 | 0 | |
| 864 | 1039 | 39 | 49 | 50 | 1 | 1 | 1 | |
| 1003 | 1013 | 12 | 11 | 10 | 1 | 0 | 1 | |
| 1003 | 1200 | 15 | 18 | 16 | 2 | 1 | 1 | |
| 1008 | 1200 | 12 | 14 | 12 | 2 | 0 | 1 | |
| B | 864 | 1200 | 34 | 24 | 24 | 1 | 1 | 1 |
| 919 | 1086 | 25 | 42 | 47 | 1 | 1 | 1 | |
| C | 865 | 1200 | 31 | 20 | 21 | 1 | 1 | 0 |
| 919 | 1051 | 63 | 22 | 43 | 1 | 0 | 0 | |
| 919 | 1055 | 57 | 19 | 36 | 2 | 2 | 0 | |
| 919 | 1060 | 51 | 25 | 36 | 1 | 0 | 0 | |
-
Part A shows the cross links, which do not distinguish between the conformations. Part B and C list cross-links, for which the mapped distances were significantly different depending on the model used (i.e., <30 Å for one model and >30 Å for another). Eight of these cross-links (shown also in Figure 6C) were detected only in the presence of ADP·BeFx and ATPγS and are listed in part C.





