DNA damage induces nuclear actin filament assembly by Formin-2 and Spire-1/2 that promotes efficient DNA repair
Figures
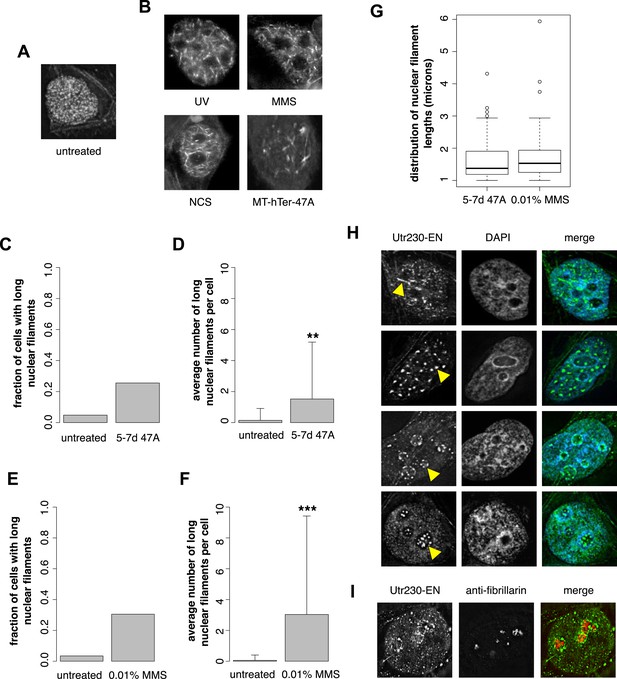
DNA damage induces nuclear actin polymerization.
(A) Localization of Utr230-EGFP-NLS (Utr230-EN) in unstressed HeLa cells. (B) Utr230-EN localization after induction of DNA damage via: 50 J/m-s UV exposure, 120-min incubation in media supplemented with 0.01% methyl methanosulfonate (MMS), 120-min incubation in media supplemented with 50 pg/ml neocarzinostatin (NCS), or 1 week post-transfection with telomere uncapping reagent MT-hTer-47A. (C) Percentage of cells with long (>1 micron) nuclear filaments after treatment with telomere uncapping reagent MT-hTer-47A (47A) for 5–7 days. N = 83–90 cells. (D) Average number of long nuclear filaments per cell after treatment with telomere uncapping reagent 47A for 5–7 days. N = 83–90 cells. (E) Percentage of cells with long (>1 micron) nuclear filaments after incubation in 0.01% MMS for 120′. N = 87–118 cells. (F) Average number of long nuclear filaments per cell incubation in 0.01% MMS for 120′. N = 87–118 cells. (G) Comparison of length distributions of long nuclear filaments after treatment with either 47A for 5–7 days or 120′ in 0.01% MMS. Open circles indicate outliers. N = 94–95 filaments. (H) Utr230-EN localization after 120 minute incubation in 0.01% MMS including several classes of nuclear filaments, listed from top to bottom: elongated nucleoplasmic, clustered, peri- and intra-nucleolar. Nuclear filaments are indicated with yellow arrows. (I) Co-localization of peri-nucleolar filaments with an antibody detecting nucleolar protein fibrillarin. Asterisks indicate p-values < 10E-3 (**) or 10E-4 (***) for all panels.
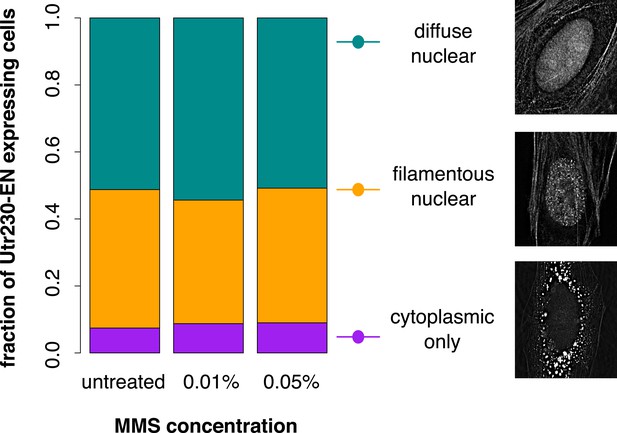
Distribution of Utr230-EN localization patterns in HeLa cells.
The subcellular distribution of Utr230-EN (diffuse nuclear and cytoplasmic, filamentous nuclear and cytoplasmic, or cytoplasmic only) in untreated cells and cells treated with 0.01% or 0.05% MMS for 120′. N = 206–311 cells.
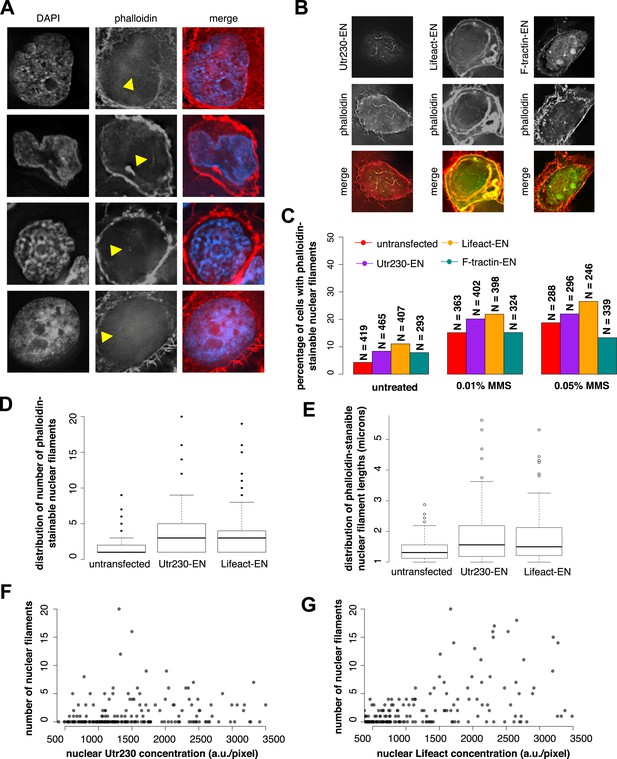
Comparison of nuclear-targeted actin reporters for detection of DNA damage-induced nuclear actin filaments.
(A) Faint detection of elongated nuclear actin filaments in the nucleoplasm and peri- and intra-nucleolar regions after 0.01% MMS treatment by phalloidin in untransfected cells. Arrows indicate locations of nuclear filaments, including elongated nucleoplasmic filaments (top two rows) and phalloidin staining of intra-nucleolar (third row) and peri-nucleolar (bottom row) structures. (B) Detection of elongated nuclear actin filaments after 0.01% MMS treatment by nuclear-targeted variants of three common live-cell actin reporters: Utr230-EN, Lifeact-EGFP-NLS (Lifeact-EN), and F-tractin-EGFP-NLS (F-tractin-EN). (C) Percentage of cells containing phalloidin-stainable nuclear filaments before and after MMS treatment within cell lines expressing actin reporters shown in (B). N values are as shown. (D) Distribution of the number of phalloidin-stainable nuclear filaments per cell after MMS treatment within cell lines expressing actin reporters shown in (B). Closed circles indicate outliers. (E) Distributions of lengths of long (>1 micron) phalloidin-stainable nuclear filaments per cell after MMS treatment within cell lines expressing actin reporters shown in (B). N = 82–100 filaments. Open circles indicate outliers. (F) Scatter plot depicting the number of Utr230-EN detected nuclear filaments per cell as a function of the concentration of Utr230-EN in the nucleus following treatment with 0.01% MMS for 120 minute. N = 206 cells. (G) Scatter plot depicting the number of Lifeact-EN detected nuclear filaments per cell as a function of the concentration of Lifeact-EN in the nucleus following treatment with 0.01% MMS for 120′. N = 214 cells.
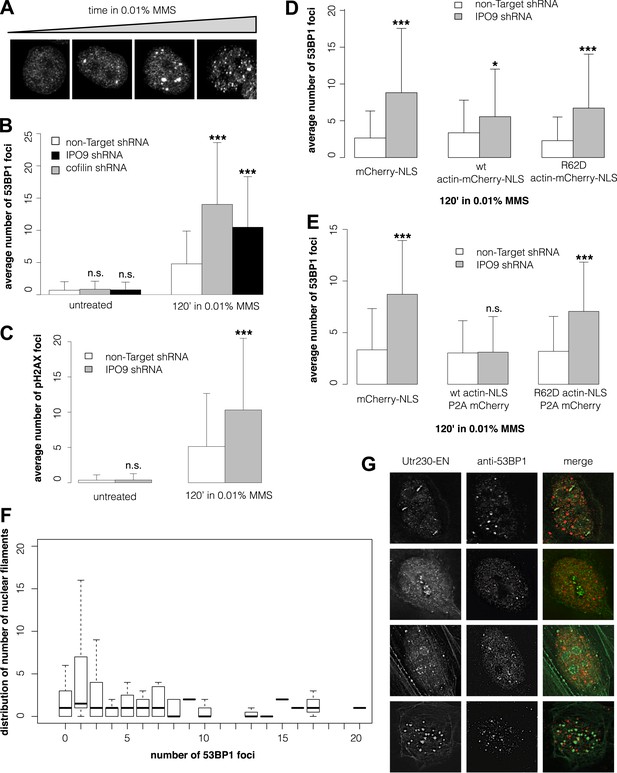
Nuclear actin polymerization is required for efficient double-strand break clearance.
(A) Double-strand break (DSB) sites detected by 53BP1 immunofluorescence after 30′, 60′, 90′, and 120′ incubations in 0.01% MMS. (B) Average number of DSB foci detected by 53BP1 immunofluorescence per cell after stable shRNA knockdown of the nuclear actin import factors, importin-9 (IPO9) or cofilin, after 0.01% MMS incubation. N = 125–172 cells per condition. (C) Average number of DSB foci detected by gamma H2AX immunofluorescence per cell after stable shRNA IPO9 knockdown following 0.01% MMS incubation. N = 182–241 cells per condition. (D) Partial rescue of control DSB foci levels after IPO9 knockdown by overexpression of wild-type actin-NLS-P2A-mCherry but not non-polymerizing R62D mutant of actin-NLS-P2A-mCherry. N = 118–150 cells per condition. (E) Full rescue of non-Target control DSB foci levels after IPO9 knockdown by overexpression of wild-type actin-NLS-P2A-mCherry but not non-polymerizing R62D mutant of actin-NLS-P2A-mCherry. N = 262–291 cells per condition. (F) Comparison of the distributions of long (>1 micron) nuclear filaments per cell and 53BP1 foci counts. N = 206 cells. (G) Co-localization assays between Utr230-EN and DSBs after 120′ incubation in 0.01% MMS. Asterisks indicate p-values < 10E-2 (*), 10E-3 (**), or 10E-4 (***) for all panels.
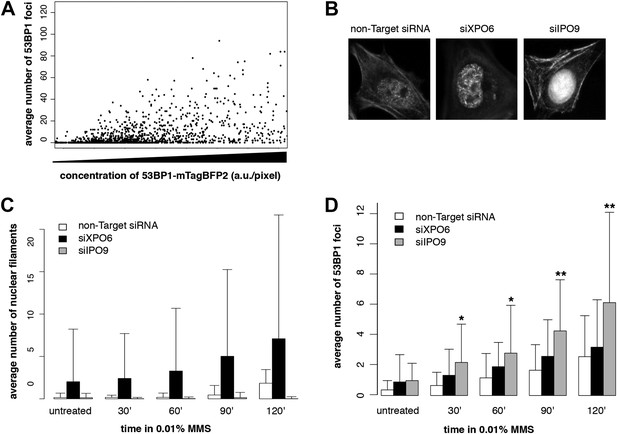
Knockdown of exportin-6 or IPO9 modulates filament formation after DNA damage.
(A) Scatter plot comparing the number of 53BP1-mTagBFP2 foci per cell and 53BP1-mTagBFP2 expression level following treatment with 0.01% MMS for 120′. N = 354 cells. (B) Localization of Utr230-EN after 0.01% MMS treatment following transient siRNA knockdown of exportin-6 (XPO6) or IPO9. (C) Quantification of elongated nuclear filament formation after increasing doses of MMS in IPO9, XPO6, and control siRNA knockdown lines. N = 126–150 cells. (D) Quantification of DSBs per cell detected by 53BP1 antibody after increasing doses of MMS in IPO9, XPO6, and control knockdown cell lines. N = 126–150 cells. Asterisks indicate p-values < 10E-2 (*) or 10E-3 (**).
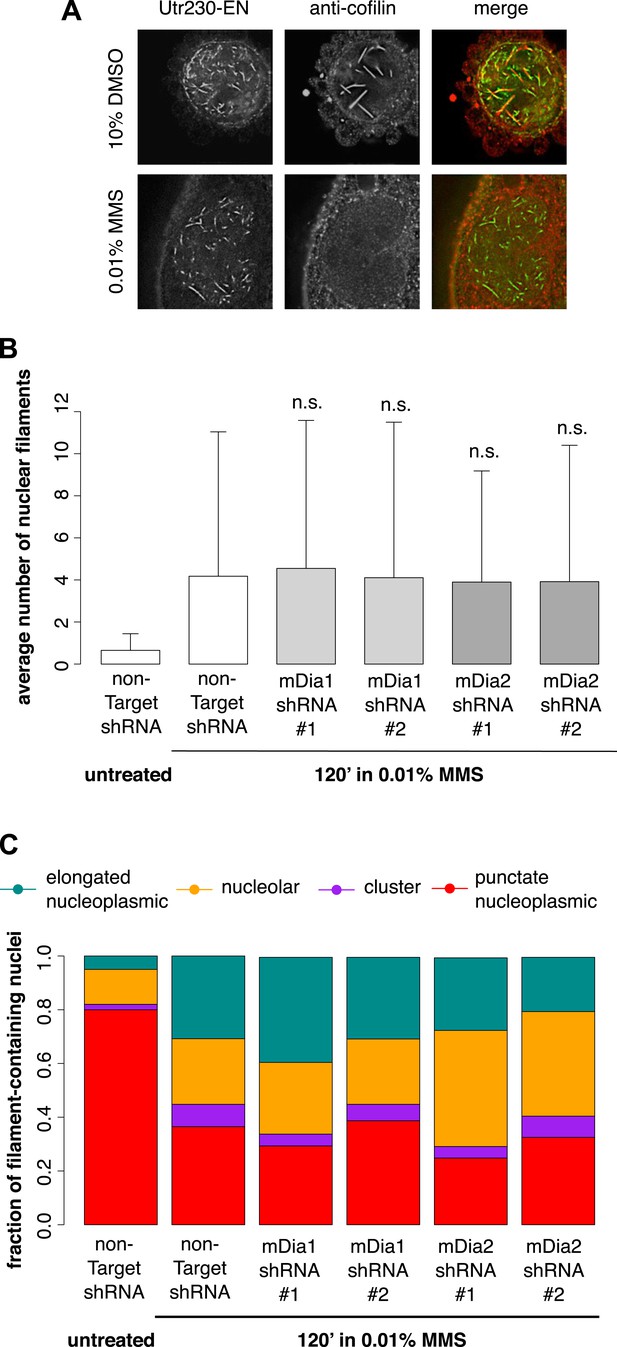
DNA damage-induced nuclear actin filaments are distinct from cofilin–actin rods and mDia1/2-generated nuclear filaments.
(A) Cofilin antibody staining in cells expressing Utr230-EN and treated with either 10% DMSO for 30′ or 0.01% MMS for 120′. (B) Average number of nuclear actin filaments per cell after 120′ 0.01% MMS treatment in cells stably expressing mDia1, mDia2, or non-Target control hairpins. N = 115–128 cells per condition. (C) Distribution of nuclear actin filament classes after 120′ 0.01% MMS treatment in cells stably expressing mDia1, mDia2, or non-Target control hairpins. N = 115–128 cells per condition.
Validation of mDia1 and mDia2 knockdown by Western blot.
Western blots in whole-cell lysate of knockdown lines generated using stably expressed Mission shRNA constructs (Sigma). The RNAi Consortium Number (TRCN) for shRNA constructs is shown. Images have been enhanced for contrast.
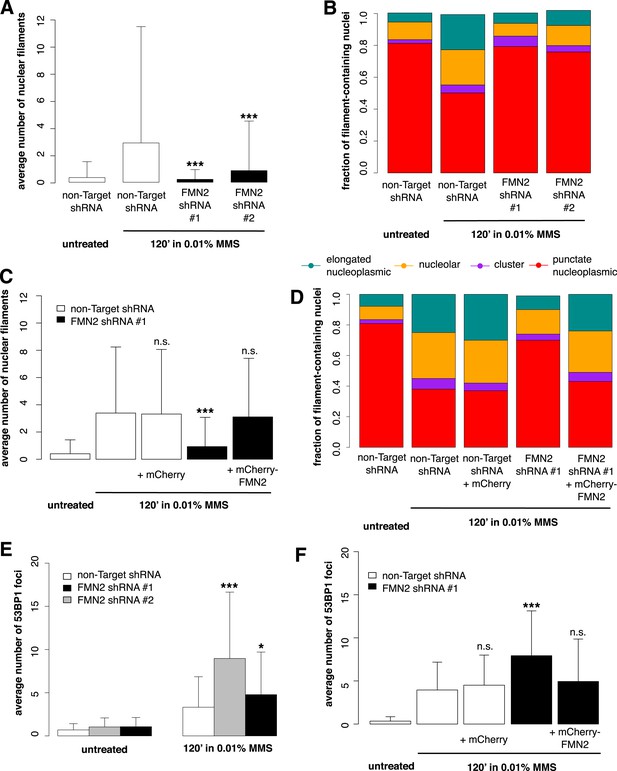
Formin-2 is required for DNA damage-induced nuclear actin polymerization.
(A) Average number of nuclear actin filaments per cell after 120′ 0.01% MMS treatment in cells stably expressing Formin-2 (FMN2) or non-Target control hairpins. N = 117–151 cells per condition. (B) Distribution of nuclear actin filament classes after 120′ 0.01% MMS treatment in cells stably expressing FMN2 or non-Target control hairpins. N = 117–151 cells per condition. (C) Rescue of FMN2 hairpin in (A) by overexpression of hairpin-resistant mutant of mCherry-FMN2. N = 105–127 cells per condition. (D) Rescue of FMN2 hairpin in (B) by overexpression of hairpin-resistant mutant of mCherry-FMN2. N = 105–127 cells per condition. (E) Knockdown of FMN2 via stably selected hairpins increases the number of DSB foci after 0.01% MMS incubation. N = 238–323 cells per condition. (F) Rescue of control DSB foci levels after knockdown of FMN2 by overexpression of hairpin-resistant mutant of mCherry-FMN2. N = 229–264 cells per condition. Asterisks indicate p-values < 10E-2 (*) or 10E-4 (***) for all panels.
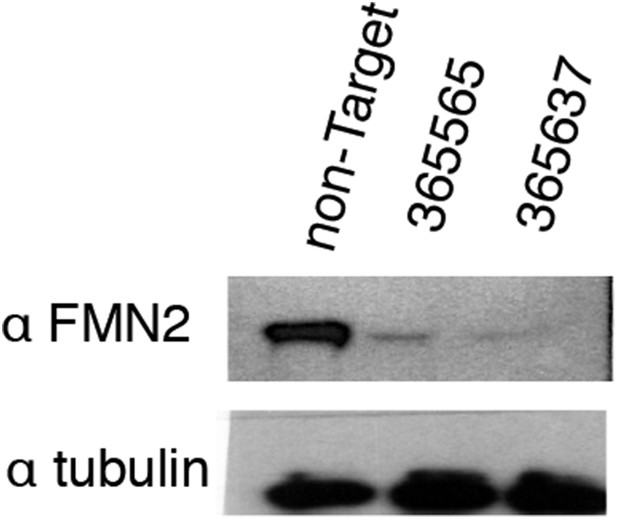
Validation of FMN2 knockdown by Western blot.
Western blots in whole-cell lysate of knockdown lines generated using stably expressed Mission shRNA constructs (Sigma). The RNAi Consortium Number (TRCN) for shRNA constructs is shown. Images have been enhanced for contrast.
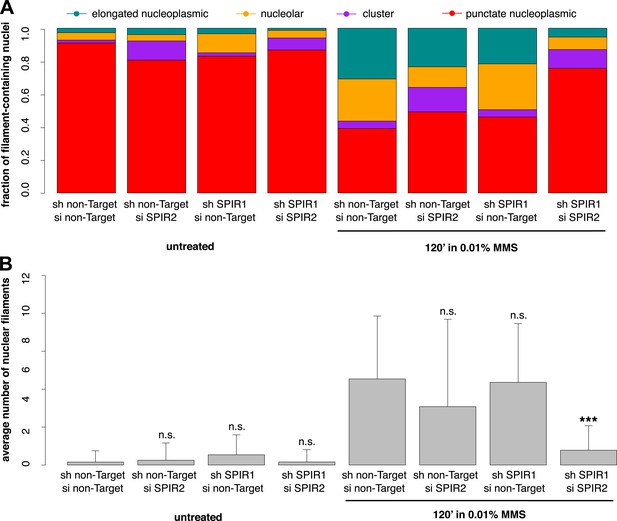
Composite knockdown of Spire-1 and Spire-2 inhibits DNA damage-induced nuclear actin assembly.
(A) Distribution of nuclear actin filament classes after 120′ 0.01% MMS treatment in cells both stably expressing either Spire-1 (SPIR1) or non-Target control hairpins and transiently transfected with either Spire2 (SPIR2) or non-Target control siRNA. N = 111–185 cells per condition. (B) Average number of nuclear actin filaments per cell for knockdown conditions described in (A). Asterisks indicate p-values < 10E-3 (**) or 10E-4 (***).
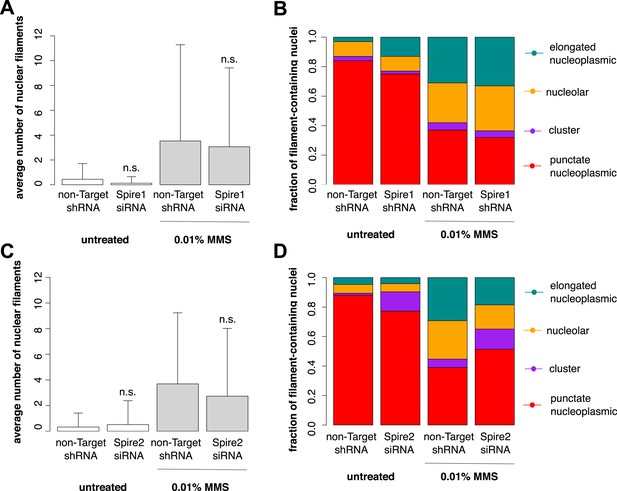
Individual knockdown of either Spire-1 or Spire2 does not affect nuclear actin filament polymerization in response to DNA damage.
(A) Average number of nuclear actin filaments per cell after 120′ 0.01% MMS treatment in cells stably expressing Spire-1 or non-Target control hairpins. N = 178–224 cells per condition. (B) Distribution of nuclear actin filament classes after 120′ 0.01% MMS treatment in cells stably expressing Spire-1 or non-Target control hairpins. N = 178–224 cells per condition. (C, D) Repeat of (A, B) for Spire-2 knockdown. N = 112–149.
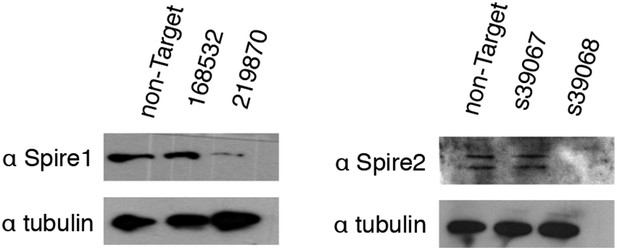
Validation of Spire-1 and Spire-2 knockdowns by Western blot.
Western blots in whole-cell lysate of knockdown lines generated using stably expressed Mission shRNA constructs (Sigma) or Silencer Select siRNAs (Life Technologies). The RNAi Consortium Number (TRCN) for shRNA constructs and product ID for siRNAs are shown. Images have been enhanced for contrast. Only knockdown reagents yielding a successful knockdown (Spire1 shRNA 219870 and Spire2 siRNA s39068) were used for analysis.
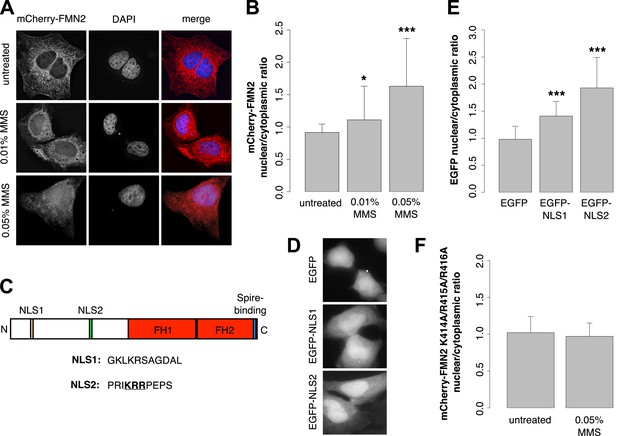
Formin-2 accumulates in the nucleus in response to DNA damage.
(A) Localization of transiently expressed mCherry-FMN2 in untreated cells and after 120′ incubation in 0.01% or 0.05% MMS. (B) Average ratio of nuclear vs cytoplasmic integrated fluorescence intensity of mCherry-FMN2 in untreated cells and after 0.01% and 0.05% MMS. N = 71–94 cells per condition. (C) Domain organization of human FMN2 including two putative nuclear localization sequence (NLS) sites identified using cNLS Mapper (Kosugi et al., 2009). (D) Localization of EGFP fused to each putative NLS. (E) Nucleocytoplasmic ratio of EGFP fused to each putative NLS. N = 140–165 cells. (F) Nucleocytoplasmic ratio of the K414A/R415A/R416A NLS2 mutant mCherry-FMN2 before and after 0.05% MMS treatment. N = 108–115 cells. Asterisks indicate p-values < 10E-3 (**) or 10E-4 (***) for all panels.
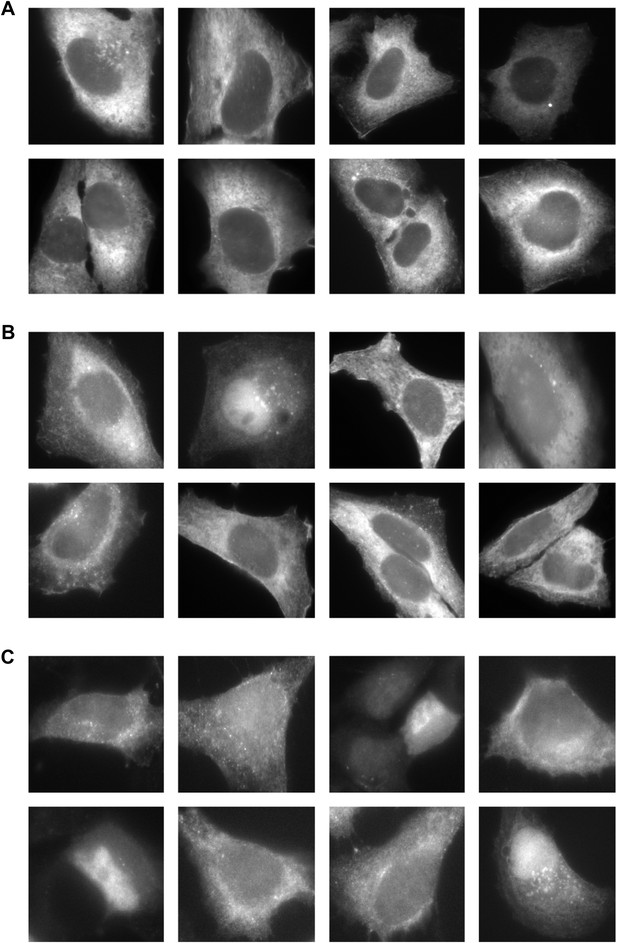
Gallery of mCherry-FMN2 localization before and after MMS treatment.
Examples of mCherry-FMN2 localization in (A) untreated cells and cells treated with (B) 0.01% MMS for 120′ or (C) 0.05% MMS for 120′.
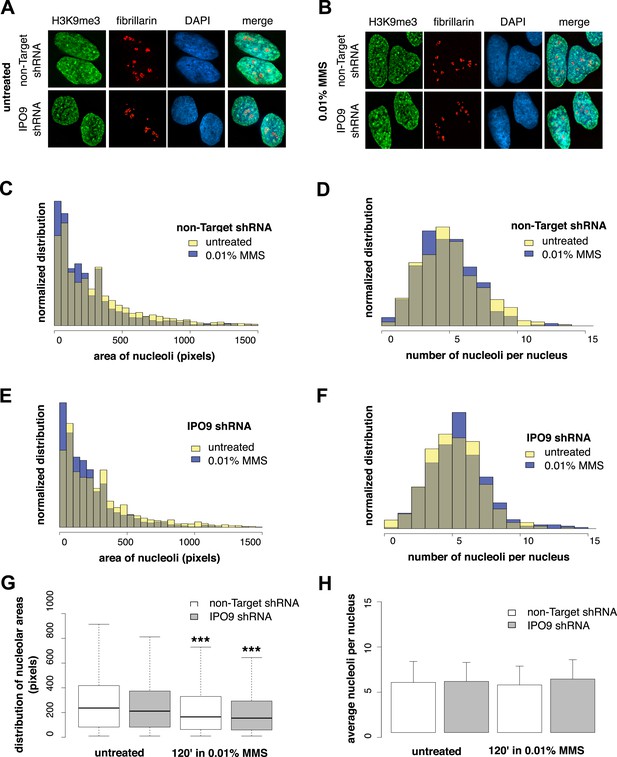
Nucleolar size decreases after DNA damage through a nuclear actin-independent pathway.
(A) Localization by immunofluorescence of heterochromatin marker H3K9me3 and nucleolar marker fibrillarin in untreated cells stably expressing non-Target and IPO9 hairpins. (B) Repeat of (A) following 0.01% MMS incubation for 120′. (C) Distribution of nucleolar areas (in pixels) before (N = 2313 nucleoli) and after 0.01% MMS incubation (N = 2299 nucleoli) in IPO9 knockdown cells. (D) Distribution of nucleolar areas (in pixels) before (N = 2094 nucleoli) and after 0.01% MMS incubation (N = 2180 nucleoli) in control cells. (E) Distribution of average numbers of nucleoli per cell before (N = 406 cells) and after 0.01% MMS incubation (N = 386 cells) in IPO9 knockdown cells. (F) Distribution of average numbers of nucleoli per cell before (N = 375 cells) and after 0.01% MMS incubation (N = 411 cells) in control cells. (G) Box plots comparing distributions shown in (C, D). Asterisks indicate p-values < 10E-4 (***). (H) Bar plots comparing averages from distributions shown in (E, F).
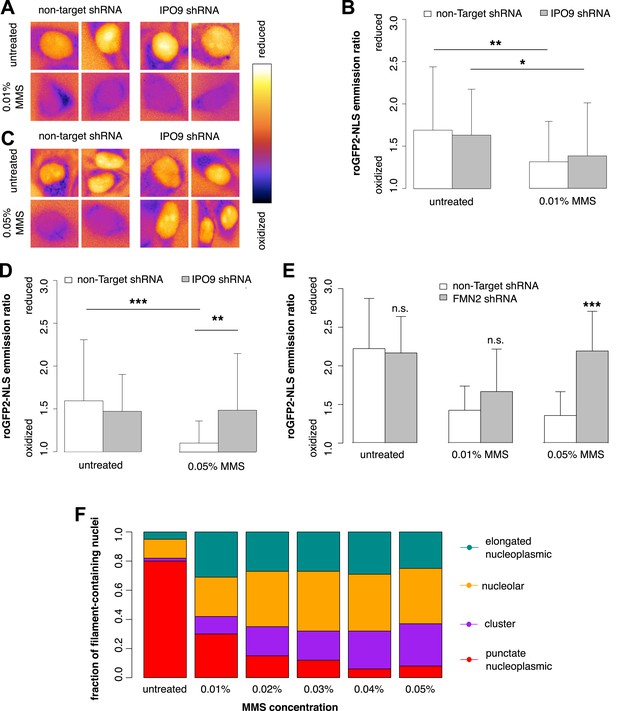
Nuclear actin is required for regulation of nuclear oxidation after acute DNA damage.
(A) False-colored images of roGFP2-NLS signal (ratio of emission intensities after 488-nm and 405-nm excitation) after 120′ in 0.01% MMS in non-Target control and IPO9 knockdown cell lines. (B) Average roGFP2-NLS signal integrated across the nuclear area in non-Target control or IPO9 hairpin lines in untreated and 0.01% MMS-treated cells. N = 108–144 cells per condition. (C) False-colored images of roGFP2-NLS signal (ratio of emission intensities after 488-nm and 405-nm excitation) after 120′ in 0.05% MMS in non-Target control and IPO9 knockdown cell lines. (D) Average roGFP2-NLS signal in nuclei of non-Target control or IPO9 hairpin lines in untreated and 0.05% MMS-treated cells. N = 106–124 cells per condition. (E) Average roGFP2-NLS signal in the nuclei of cells stably expressing non-Target or FMN2 shRNA in untreated cells and cells treated with 0.01% or 0.05% MMS for 120′. N = 109–162 cells. (F) Distribution of nuclear actin filament classes after incubation in MMS concentrations from 0.01% to 0.05% for 120′. N = 95–159 cells per condition. Asterisks indicate p-values < 10E-3 (**) or 10E-4 (***) for all panels.
Additional files
-
Source code 1
Custom ImageJ macros.
- https://doi.org/10.7554/eLife.07735.019





