Quantitative perturbation-based analysis of gene expression predicts enhancer activity in early Drosophila embryo
Figures
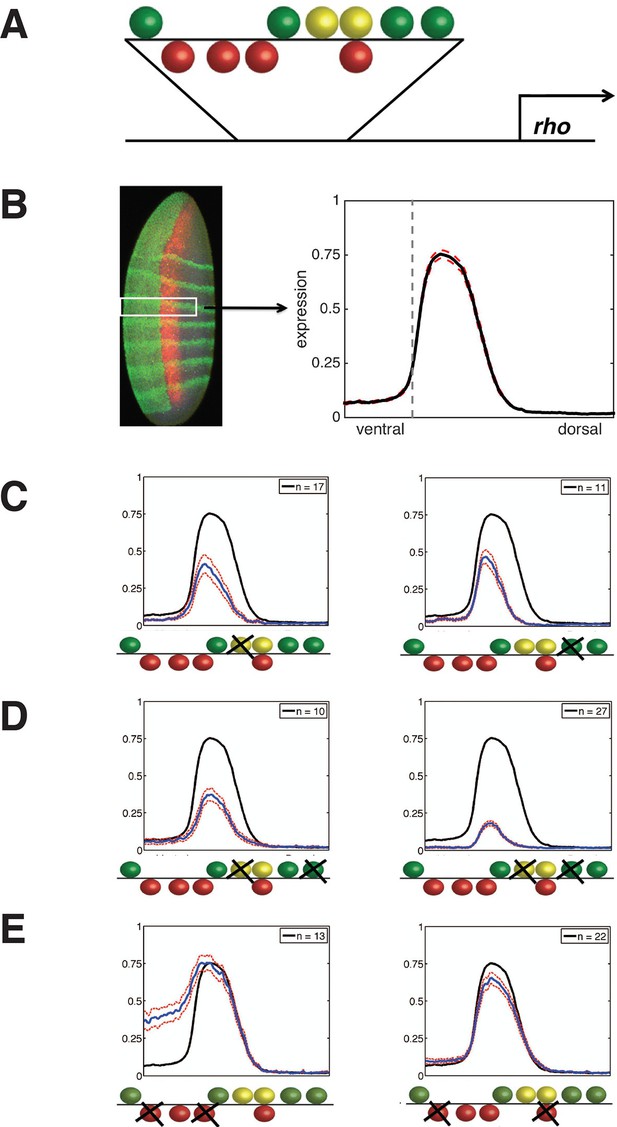
Experimental deep perturbation analysis of a neuroectoderm-specific enhancer.
(A) rhomboid (rho) enhancer with footprinted Dorsal activator sites shown in green, Twist activator sites in yellow, and Snail repressor sites in red. (B) Drosophila embryo stained to measure rho reporter readout in the neuroectoderm with quantitative expression measured using confocal laser scanning microscopy. The embryo is oriented anterior up and dorsal to the right. rho-lacZ mRNA is shown in red, seven even-skipped mRNA stripes in green, and snail mRNA in the mesoderm (left) also in green. Average expression after normalization over embryos containing the wild-type enhancer in the region of interest (white box) is plotted in black at right; dashed red line indicates standard error; vertical dotted line indicating dorsal-most boundary of sna expression. (C) Effects on expression of removal of single activator sites – expression levels decrease by a similar extent when either Dorsal or Twist activator sites are removed. (D) Effects of double activator site mutation – expression levels are variably impacted depending on which pairs of sites are affected. (E) Effects on expression of removal of pairs of Snail repressor sites, leading to partial depression in ventral regions, or no derepression. For panels C–E, solid black lines indicate wild-type enhancer expression; solid blue lines indicate expression of the tested rho enhancer; dashed red lines indicate standard error; n = number of embryos imaged for the given construct shown (in top right of each panel).
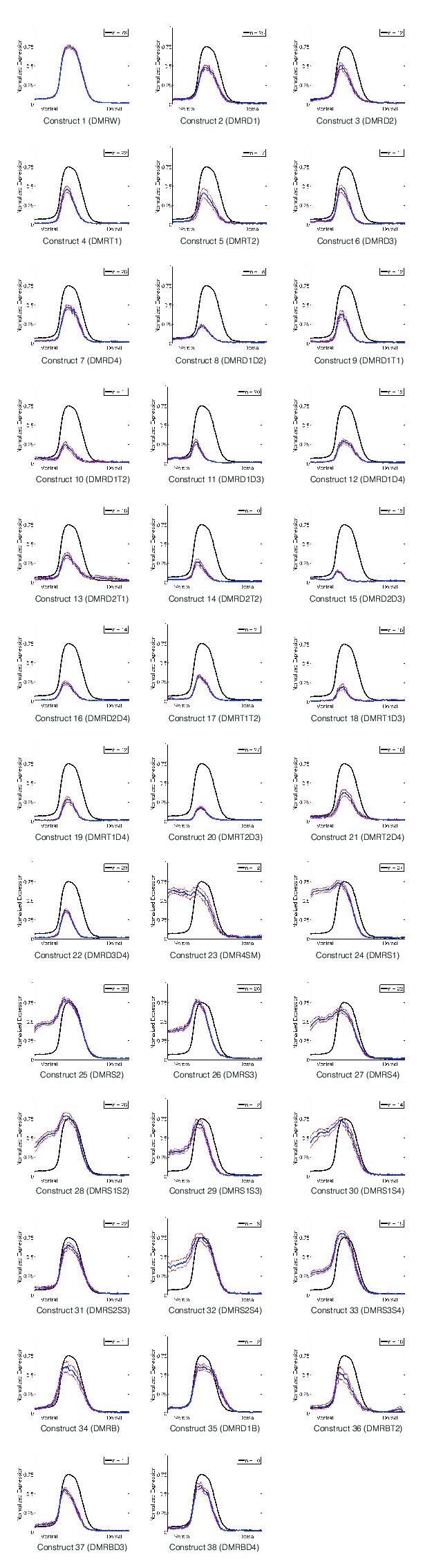
Quantitative perturbation analysis of rho neuroectodermal enhancer.
Averaged quantitative expression for each of the 38 variations of the rho enhancer that were tested; standard error of measurements indicated by gray lines surrounding mean values. Number of embryos imaged for each construct noted in corner (n).
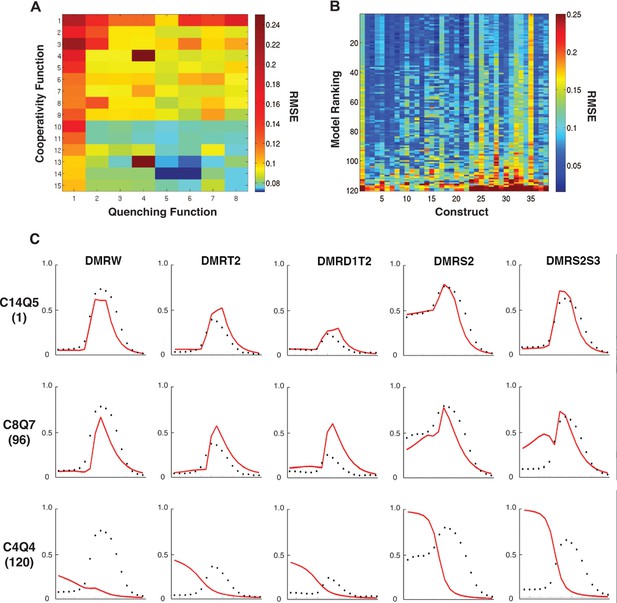
Performance of 120 thermodynamic models.
(A) Heat map representation of results of thermodynamic modeling of rho perturbation dataset; the performance of each model is represented by a rectangle. Models were globally fit using Root Mean Square Error (RMSE), scale shown at right. Each model employs a distinct repression (quenching) scheme represented on horizontal axis, and a protein-protein cooperativity scheme on vertical axis. (B) Construct-by-construct performance ranking of 120 thermodynamic models ranked top to bottom. Vertical features are indicative of specific constructs that showed overall better or worse fit. (C) Examples of fitting of individual constructs by models ranking high (C14Q5; 1st overall), medium (C8Q7; 96th overall), or low (C4Q4; 120th overall). Black points indicate measured average mRNA levels; red lines are model predictions. The high-ranking model correctly predicts both low expression (repression) in mesoderm and activation in neuroectoderm, medium model exhibits modest deviations in prediction of both repression and activation, and the low-ranked model exhibits particularly poor fits for repression activity.
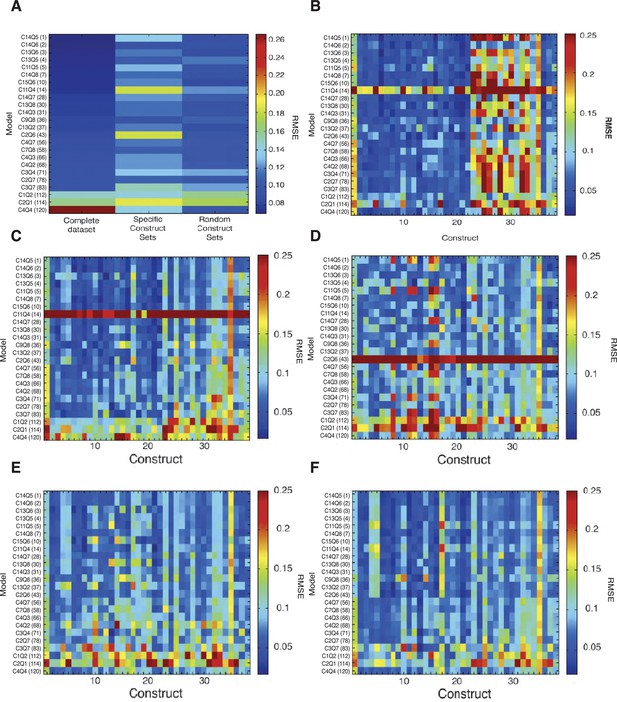
Cross validation of thermodynamic models.
(A) Random five-fold cross validation was performed on 24 models as described in Materials and Methods to test for overfitting. Compared to fitting on the complete data set, most models showed only a modest decrease in performance in predicting left out sets (left column, complete data set, right column, modeling on randomly selected partial data sets). In contrast, selective removal of specific data sets corresponding to one type of mutagenic perturbation (all constructs with mutation to Snail sites, all constructs with mutations to Dorsal sites, etc.) led to parameter sets that were generally significantly less able to fit left-out data (central column). The comprehensive perturbation of the different types of motifs on the enhancers was thus an essential aspect of good model fitting. The low-performing model C4Q4 actually performed better during cross-validation, likely because the parameters determined from the original run do not represent a global minimum. (B) Construct-specific RMSE scores illustrate impact of specific portions of the data set on model performance. Here, predictions for 38 constructs are plotted for the 24 models that were fit on a data set lacking all Snail mutant constructs. The performance is specifically diminished for constructs 23–33, which represent Snail mutant enhancers (compare to Figure 2B). (C) Removal of bHLH site perturbation constructs changes little in overall fitting. (D) The removal of constructs with Dorsal site mutations is associated with poorer performance across a spectrum of models on constructs that represent Dorsal site mutants (columns 8, 11–12, 15–16). (E) Similarly, omission of mutant constructs with perturbed Dorsal and Twist sites degrades performance especially on constructs 4–5, 9–10, 14, 17–21 that represent Dorsal+Twist mutations. (F) Removal of Twist alone mutants especially impacts fitting on construct 17, the Twist1+2 mutant construct. Weak performance of C11Q4 and C2Q6 across the spectrum in B, C, and D represents suboptimal parameter estimation by the genetic algorithm specifically in these runs.
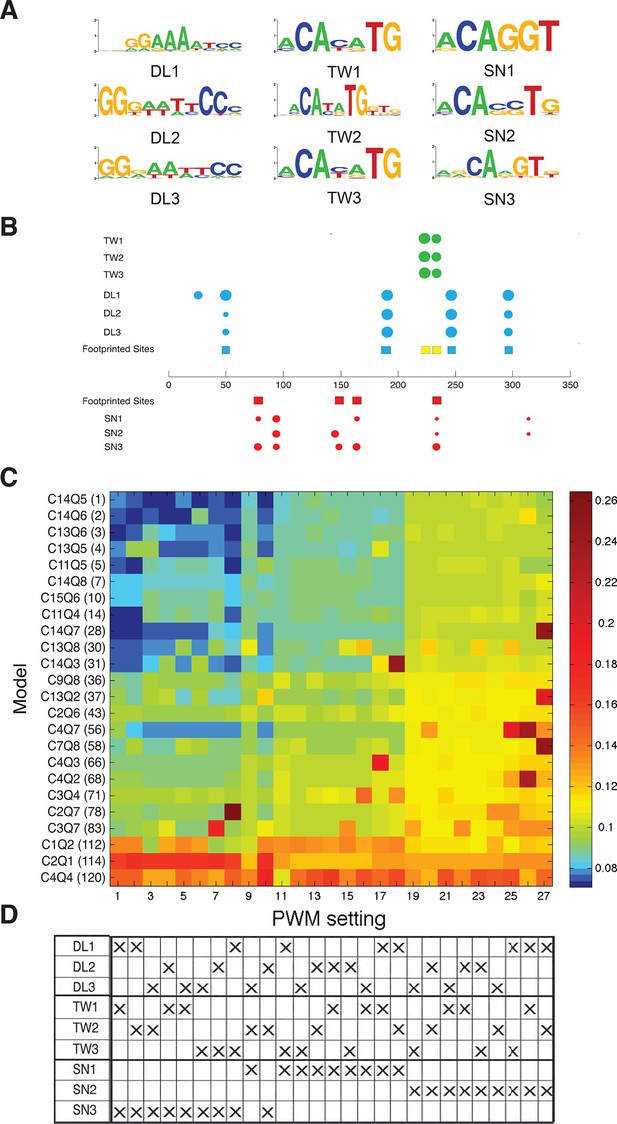
Effect of PWM settings on model performance.
(A) Three versions of each binding site were considered as discussed in Materials and methods, drawing from previously reported in vitro or in vivo binding studies. (B) The correspondence between previously identified bindings sites from DNAseI footprinting studies (colored squares) and predicted binding sites using the diverse PWMs (circles) is superimposed on the structure of the rho enhancer. The size of the circles indicates the significance of the match using the particular PWM for that factor; Twist sites in green, Dorsal in blue, and Snail in red. (C) Heatmap showing global performance of 24 selected models using 27 different PWM combinations for Dorsal, Twist and Snail. Results were clustered by performance; columns of PWM settings were arranged with lowest average RMSE values to the left, then rows of models were arranged with lowest global RMSE towards the top. (D) PWMs used for different settings are denoted by ‘X’s; Snail3 PWM (used in the rest of this study) shows strong correlation with best model fitting.
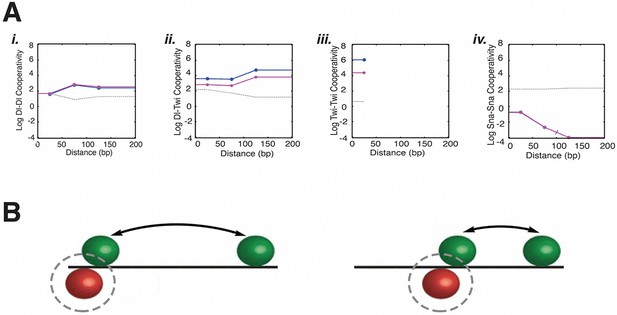
Estimated model parameters suggest biological properties of long-distance activator-activator cooperativity and weak repressor cooperativity.
(A) Representative estimated cooperativity parameters for Dorsal and Twist transcriptional activators show a trend of high levels of cooperativity with little distance dependence, consistent with indirect, long-range effects i. Dorsal-Dorsal, ii. Dorsal-Twist, iii. Twist-Twist, and iv. Snail-Snail cooperativity values of model C13Q3. Low values estimated for repressor cooperativity are consistent with weak interactions between Snail proteins. One of the five runs for fitting this model had significantly higher RMSE than the other four; the corresponding parameters are indicated by a gray line. Other runs produced similar RMSE and parameters (magenta and blue). (B) Illustration of how global long-range cooperative interactions between activators permit relaxed constraints on binding site arrangement, while distance-dependent repressor positioning anchors Snail sites to activator sites.
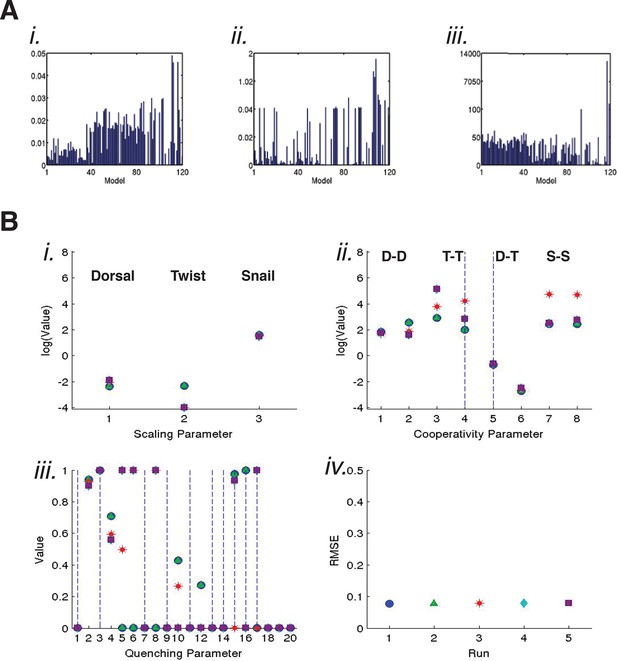
Global trends observed for parameter values and evidence of compensation.
(A) Scaling factors (related to estimated transcription factor 'potency') identified by 120 models for i. Dorsal, ii. Twist, and iii. Snail. Models ranking 1–34 had similar, lower scaling factors for Dorsal, compared to relatively higher values for models ranking 35–86. The scaling factors for the lowest ranked models 87–120 varied widely. No such trends were observed for better or worse performing models with respect to Twist scaling factors; significantly, there are only two Twist sites on the minimal rho enhancer, and thus fewer perturbations to constrain these values. Snail scaling factor values were clustered for the top 40 models, and then showed greater variation for lower performing models. For each model, the scaling factor is shown for the parameter estimation run (of five total) with the lowest RMSE. (B) Parameters estimated for model C11Q8 for i. scaling factors, ii. homotypic and heterotypic cooperativity, and iii. quenching parameters representing action of Snail on Dorsal and Twist. Global parameter estimation was carried out five times, with overall RMSE shown iv.. Although global fits were very similar, the parameter values vary, and showed evidence of compensation. In particular, the scaling factor for Twist obtained in the first and second runs (green triangle and blue square) is higher than the values obtained in the other runs. This higher scaling factor parameter is associated with a markedly lower Twist-Twist homotypic cooperativity parameter, shown in position 3 (the only interval for this model that is filled; dashed line in lane 4 signifies that there are no Twist sites at the spacing represented by this interval). The trade-off between overall activity and cooperativity is likely to account for much of the second order effects noted for many models (Figure 4—figure supplement 2). In iii., Snail quenching parameters for action on Dorsal (1–10) and Twist (11–20) show generally lower values for more distant 'bins', consistent with a short-range of action.
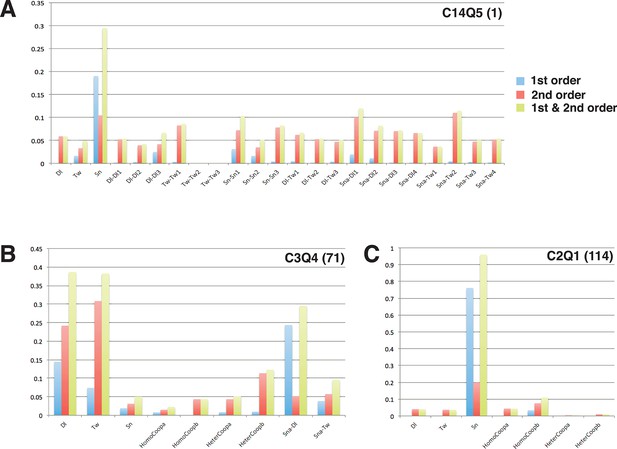
Model sensitivity analysis.
The 24 selected models used for PWM and cross validation were examined for first- and second-order relative sensitivity (complete results in Figure 4—figure supplement 2; Figure 4—figure supplement 2—source data 1). Here we show high-scoring C14Q5 model (A), as well as lower-scoring C3Q4 and C2Q1 models (B, C). As shown with these three examples, most parameters exhibited much higher second-order than first-order effects, indicating that compensatory relationships among parameters are predominant. Even with such compensation, certain types of parameters (e.g. relatively high activator cooperativities) are preferred (also seen in Figure 4—figure supplement 1). Model structures influenced the first- and second-order sensitivities; models using quenching schemes Q2-4 exhibited a significant degree of first order sensitivity for the Dorsal scaling factor (B), while almost every other model combination showed a significant extent of first-order sensitivity for the Snail scaling factor (A, C). Overall sensitivities, first- and second-order combined, were dominated in most models by those for the Snail scaling factor, consistent with the significant impact the particular Snail PWM had on modeling performance (see Figure 3).
-
Figure 4—figure supplement 2—source data 1
Sensitivity analysis.
Sensitivity analysis for each model including first- and second-order sensitivity of parameters and plots of these values.
- https://doi.org/10.7554/eLife.08445.012
Application of rho cis-regulatory 'grammar' to heterologous regulatory elements.
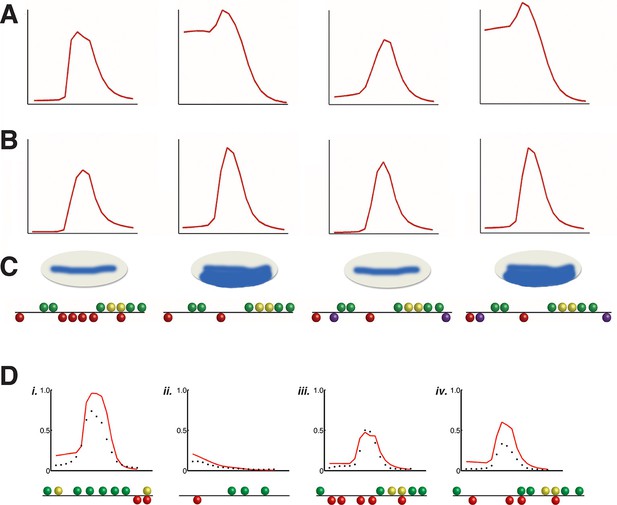
Discovery of short-range repression from modeling of rho perturbation dataset. (A) High ranking model C13Q6 (3rd overall) correctly predicts distance-dependent significance of Snail sites within enhancer element; specifically, the third but not fourth construct positioned ectopic Snail binding sites (purple balls) close enough to Dorsal 1 and Dorsal 4 motifs to repress the expression in ventral-most parts of the embryo. (B) lower ranking model C2Q7 (78th overall) correctly predicts activation potential, but fails to detect distance effects impacting repression by Snail. (C) Shows cartoons of the qualitative expression found by Gray and colleagues and the reporter genes tested in embryos (Gray et al., 1994). (D) Correctly predicted expression patterns of cis-regulatory elements i. D. melanogaster vnd enhancer, ii. D. melanogaster twi proximal element, iii. D. erecta rho enhancer, and iv. D. ananassae rho enhancer. X-axis denotes distance from ventral end of the embryo, and Y-axis denotes expression values. Predictions of model C14Q5 (ranked 1st overall) are shown as red lines; measured average transgene expression in black points.
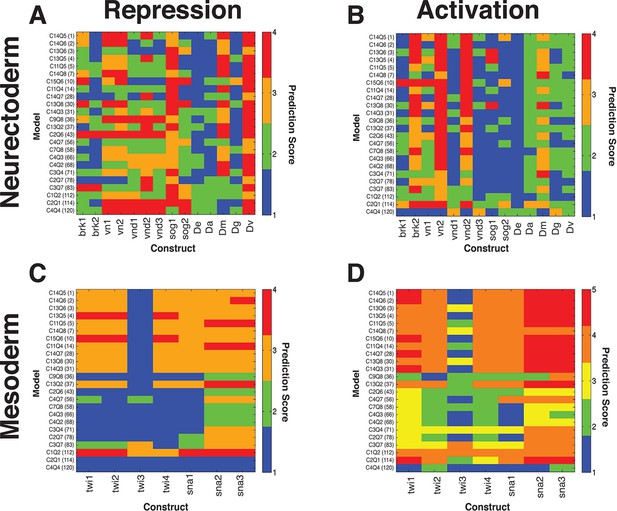
Thermodynamic models predict quantitative outputs of additional, non-rho enhancers in D. melanogaster and putative rho enhancers from related drosophilids.
(A, B) neuroectodermal elements scored for repression by Snail and activation by Dorsal and Twist, respectively. (C, D) mesodermal elements scored for lack of repression by Snail and preferential activation in mesoderm, respectively. Lowest ranked models performed poorly for correct prediction of Snail activity on neuroectodermal enhancers, but were also not liable to make a false positive call on Snail activity for mesodermal elements (A, C). The D. erecta rho element was correctly called by most models, while some sog, vn, vnd, and brk constructs were less well fit by most models, indicating that there are likely enhancer-specific features of these elements that are not sampled by rho variations. For mesodermal elements derived from snail and twist proximal regions, those models that correctly scored Snail activity as low were able to produce more 'mesoderm-specific' type patterns. No model scored consistently highly across all constructs, but many performed well in a complementary fashion, suggesting that individual models are not overfit in identical manners. Activity of regulatory elements was experimentally measured in transgenic D. melanogaster, and output compared with the predicted expression from 24 models thermodynamic models fit on D. melanogaster rho expression. Fitting for predicted vs. measured expression was assessed using quantitative measures for correct expression as described in Materials and Methods. Lower scores represent better fits. D.erecta, D.ananassae, D.mojavensis, D.grimshawi, and D.virilis putative rho elements were assayed (sequences indicated in Supplementary file 1).
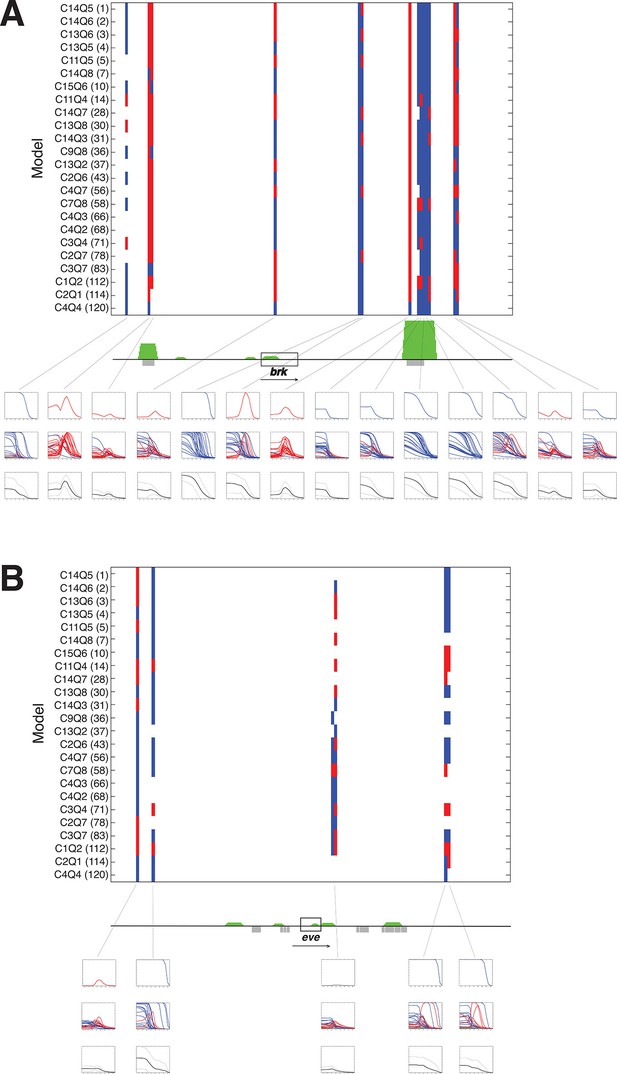
A panel of models fit on rho enhancer predicts additional neuroectodermal enhancer elements.
(A) Predicted expression activity of a genomic region (36 kb) flanking the neurectodermal brk gene, using a panel of 24 models, including 21 higher and three lower performing models. Blue marks indicate predicted mesodermal pattern, red neuroectodermal. The transcribed area for brk is indicated by the black rectangle; green peaks indicate in vivo binding for Dorsal, Twist, and Snail; previously mapped embryonic enhancers are indicated by gray rectangles. Rows of plots below the gene show expression patterns of: first row – top model, second row – composite of all 24 model predictions, third row – average of all 24 model predictions in solid black with standard deviation from the mean shown above and below in dotted black lines. Note that the average plot is similar to a plot of a weighted average using RMSEs or AICs to weight each model’s output. (B) Similar scan of the even-skipped locus (32 kb), which is not regulated by Dorsal, and where few areas are consistently identified as potential Dorsal/Twist/Snail enhancers.
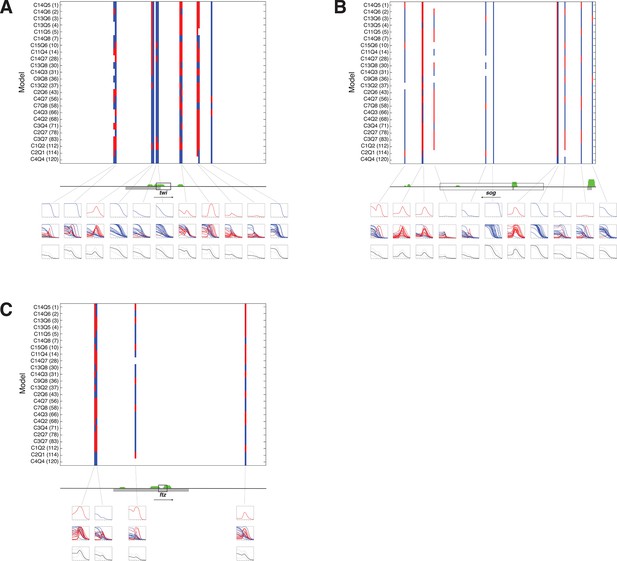
A panel of models fit on rho enhancer predicts additional neuroectodermal enhancer elements around sog and twi, with lower scores around ftz control region.
https://doi.org/10.7554/eLife.08445.016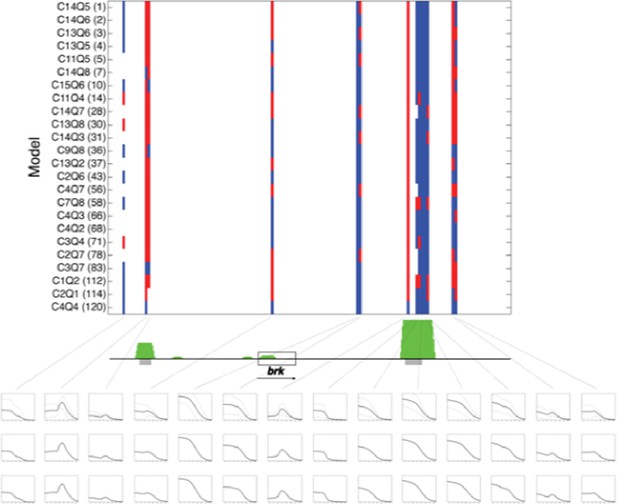
Predictions of twenty four rho-fit thermodynamic models for the brinker genomic region.
Model structure indicated by nomenclature indicating cooperativity/quenching schemes used, and overall ranking in fitting for rho training set. Red indicates neuroectodermal-like pattern; blue mesodermal-like pattern prediction. brk transcription unit indicated by gray box. Green peaks indicate ChIP-seq measured binding of regulators Twist, Snail and Dorsal. Gray bars indicate experimentally validated enhancers. Boxes below indicate (top to bottom) simple averages of ensemble of models with standard deviation, RMSE weighted averages, and AIC weighted averages. For these predictions, the simple average closely resembled the other two methods..
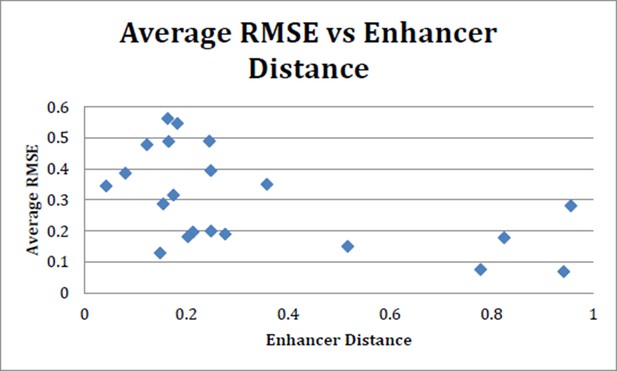
Average RMSE (suite of 24 models) vs enhancer distance as calculated using N2 method.
There is no strong trend toward increased performance (lower average RMSE) with more closely related enhancers. N2 method run using code provided in Göke J et al. (Bioinformatics, 2012).
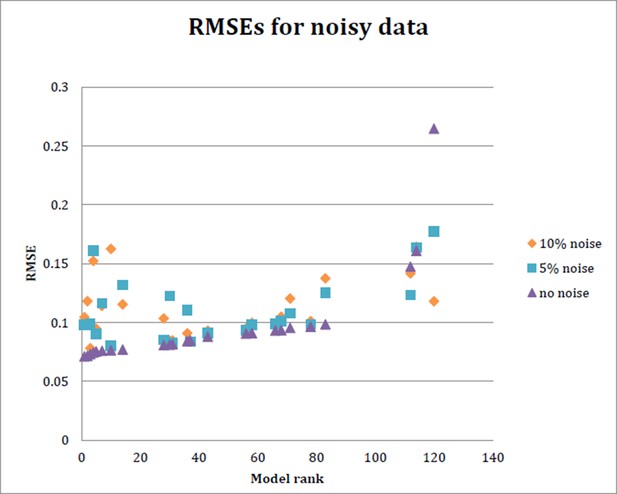
RMSE for each of 24 models run with data simulating 5% noise and 10% noise.
Ten datasets were generated to randomly introduce 5% or 10% noise, respectively, into the input data. The model was then fit on each of these noisy datasets as before, and the average RMSE obtained from the 10 datasets at 5% noise and 10% noise is plotted here, as well as the RMSE obtained with initial model fitting..
Tables
Parameters in each model. Number of parameters in cooperativity and quenching model combinations. 3 scaling factor parameters for Dorsal, Twist, and Snail are included in all models. Column 1 contains the nomenclature and number of parameters (in parentheses) for all 15 cooperativity models. Columns 2–9 in Row 2 contain nomenclature and number of parameters (in parentheses) for 8 quenching models. Each cooperativity and quenching scheme is described in the materials and methods section. The parameters being fitted for continuous functions are clearly laid out in this section. The following example illustrates the parameters in binned cooperativity and quenching functions. Model C14Q5: Parameters 1–3 are scaling factors for Dorsal, Twist, and Snail respectively. Parameters 4–15 reflect cooperativity (separate parameters existing for each type of protein-protein interaction)– parameters 4–6 represent Dorsal-Dorsal cooperativity (at 1–60 bp, 61–120 bp, and 121+ bp between bound Dorsal proteins), likewise parameters 7–9 represent Twist-Twist cooperativity, parameters 10–12 represent Dorsal-Twist cooperativity, and parameters 13–15 represent Snail-Snail cooperativity. Paramters 16–23 reflect quenching – parameters 16–19 represent Snail quenching Dorsal (at 1–25 bp, 26–50 bp, 51–75 bp, and 76+ bp between bound proteins), likewise parameters 20–23 represent Snail quenching Twist.
| Quenching Model | ||||||||
|---|---|---|---|---|---|---|---|---|
| Cooperativity Model | Q1 – MSB (0) | Q2 – Linear (2) | Q3 – Logistic (2) | Q4 – Gaussian (2) | Q5 – Binned 4_25 (8) | Q6 – Binned 4_35 (8) | Q7 – Binned 4_45 (8) | Q8 – Binned 10_10 (20) |
| C1 – Linear (4) | 7 | 9 | 9 | 9 | 15 | 15 | 15 | 27 |
| C2 – Logistic (4) | 7 | 9 | 9 | 9 | 15 | 15 | 15 | 27 |
| C3 – Gaussian (4) | 7 | 9 | 9 | 9 | 15 | 15 | 15 | 27 |
| C4 – Binned 2_25 (4) | 7 | 9 | 9 | 9 | 15 | 15 | 15 | 27 |
| C5 – Binned 2_50 (4) | 7 | 9 | 9 | 9 | 15 | 15 | 15 | 27 |
| C6 – Binned 2_75 (4) | 7 | 9 | 9 | 9 | 15 | 15 | 15 | 27 |
| C7 – Binned 3_50 (6) | 9 | 11 | 11 | 11 | 17 | 17 | 17 | 29 |
| C8 – Binned 3_60 (6) | 9 | 11 | 11 | 11 | 17 | 17 | 17 | 29 |
| C9 – Binned 3_70 (6) | 9 | 11 | 11 | 11 | 17 | 17 | 17 | 29 |
| C10 – Protein Binned 2_25 (8) | 11 | 13 | 13 | 13 | 19 | 19 | 19 | 31 |
| C11 – Protein Binned 2_50 (8) | 11 | 13 | 13 | 13 | 19 | 19 | 19 | 31 |
| C12 – Protein Binned 2_75 (8) | 11 | 13 | 13 | 13 | 19 | 19 | 19 | 31 |
| C13 – Protein Binned 3_50 (12) | 15 | 17 | 17 | 17 | 23 | 23 | 23 | 35 |
| C14 – Protein Binned 3_60 (12) | 15 | 17 | 17 | 17 | 23 | 23 | 23 | 35 |
| C15 – Protein Binned 3_70 (12) | 15 | 17 | 17 | 17 | 23 | 23 | 23 | 35 |
Additional files
-
Supplementary file 1
Description and sequence information for constructs List of all constructs with numbers, names, and sequences.
Restriction sites are capitals, with red capital letters indicating mutated binding site sequences.
- https://doi.org/10.7554/eLife.08445.017
-
Supplementary file 2
Expression data for constructs Quantification of signal intensity along the DV axis of the embryo with standard error for each construct.
Sheet 2 has the 17 data points used for fitting on each construct.
- https://doi.org/10.7554/eLife.08445.018
-
Supplementary file 3
Parameters for all model runs List of parameters for all five runs of each model.
- https://doi.org/10.7554/eLife.08445.019





