The RhoGAP activity of CYK-4/MgcRacGAP functions non-canonically by promoting RhoA activation during cytokinesis
Figures
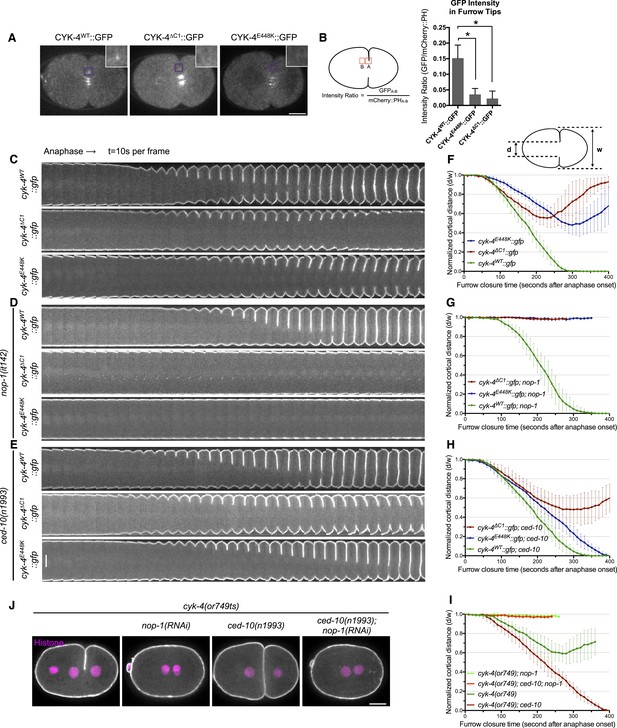
CYK-4-dependent membrane binding promotes furrow ingression.
(A) CYK-4 accumulates on the plasma membrane. Membrane accumulation is observed on ingressing cleavage furrows (boxed regions). CYK-4 membrane accumulation requires the C1 domain and is compromised by the E448K substitution in the cyk-4(or749ts) allele. (B) Membrane accumulation of CYK-4 variants. The accumulation was quantified as a ratio of the accumulation of CYK-4::GFP/mCherry::PH at the furrow tip as depicted in the schematic; the mean intensity ±s.e.m. are plotted. (N = 8–12 embryos *p < 0.05, one way ANOVA followed by Tukey multiple comparison). (C) Deletion of the C1 domain and the E448K substitution impair cleavage furrow ingression. Kymographs generated from time-lapse recordings of the equatorial region of embryos of the indicated genotypes expressing the membrane marker mCherry::PH. Kymographs begin at anaphase onset. (D) Deletion of the C1 domain and the E448K substitution abrogate centralspindlin-dependent furrow ingression. The progression of cytokinesis was assessed in embryos expressing CYK-4 variants in combination with a loss of function mutation in nop-1. (E) Mutation of CED-10/Rac1 slightly increases furrow ingression in CYK-4∆C1 embryos and allows complete, albeit delayed, furrow ingression in CYK-4E448K embryos. The progression of cytokinesis was assessed in embryos expressing CYK-4 variants in combination with a loss of function mutation in ced-10. (F–I) Quantification of furrow ingression rates in embryos of the indicated genotypes. Representative examples are shown in the accompanying kymographs. (N = 8–12 embryos; error bars, 95% confidence intervals). (J) The ability of ced-10/Rac1 to rescue cytokinesis in CYK-4E448K embryos requires the NOP-1 pathway for furrow ingression. Images shown reflect the maximal extent of furrow ingression in embryos of the indicated genotypes expressing the membrane marker mCherry::PH. Unless otherwise specified, all scale bars in all figures are 10 µm.
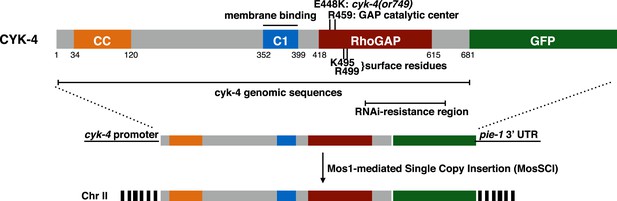
CYK-4 structure, mutations, and methods of transgene integration.
Schematic depiction of the domain organization of CYK-4, the variants tested, and the method for single copy transgene integration at a defined locus.
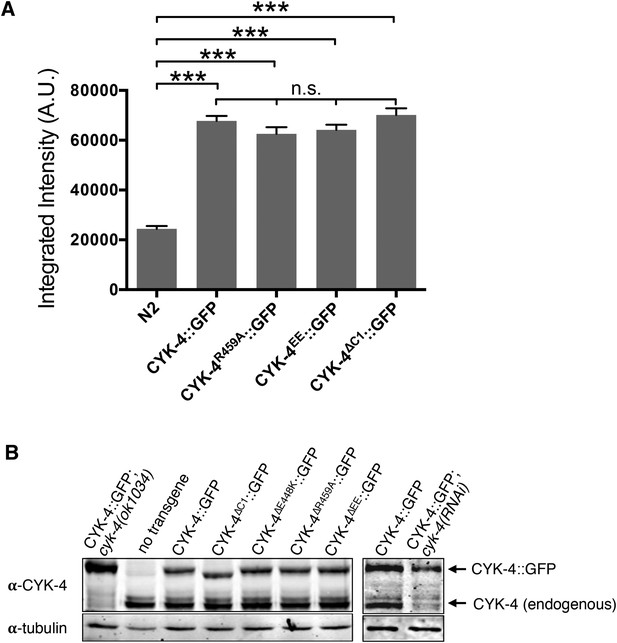
Transgene expression levels.
(A) Quantification of the expression of CYK-4::GFP transgenes in embryos. Embryos were assayed in the first cell division and the total fluorescence intensity was measured. (N ≥ 8 embryos; error bars, s.e.m; ***p < 0.001; n.s., not significant, by one way ANOVA followed by Tukey multiple comparison). (B) Quantification of the expression of CYK-4::GFP transgenes. Total worm lysates were prepared from adult worms, standardized for total protein concentration and blotted with an anti-CYK-4 antibody. In the case of the RNAi experiment, 120 gravid adults worms were treated with cyk-4(RNAi) or control treated and subjected to western blotting.
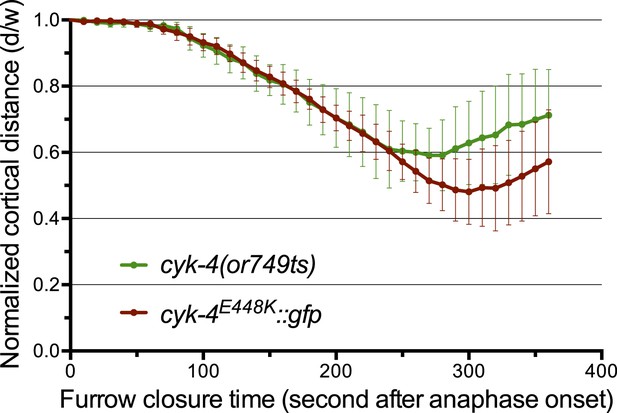
The CYK-4E448K::GFP transgene combined with cyk-4(RNAi) closely phenocopies cyk-4(or749ts).
Quantification of furrow ingression rates in embryos of the indicated genotypes. Results are quantified as described in Figure 1G.
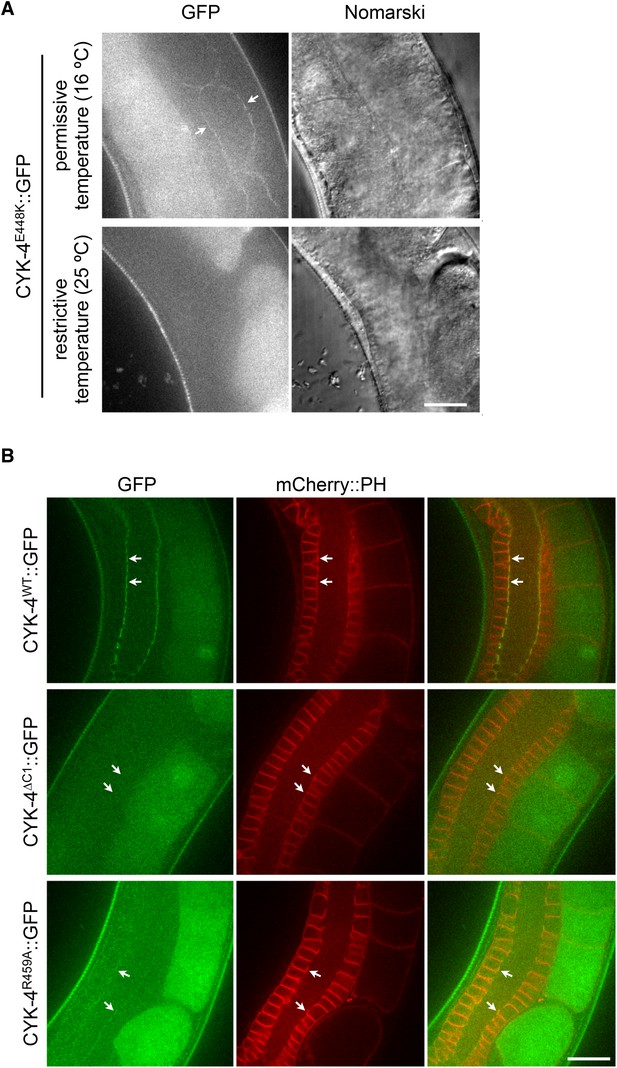
Assay for localization of CYK-4 variants to the membrane of the gonad.
(A) CYK-4E448K exhibits temperature sensitive binding to the incomplete membranes of the syncytial hermaphrodite gonad. Animals in which both endogenous and the GFP-tagged transgene contain the E448K substitution were imaged either immediately after removal from a 16°C incubator or after 60 min at 25°C. At the permissive temperature, CYK-4E448K binds to the gonad membrane, but not at the restrictive temperature. (B) The localization of CYK-4WT, CYK-4∆C1, and CYK-4R459A was analyzed in hermaphrodite gonads. mCherry::PH was used to mark the membranes in the syncytial gonad. Whereas both CYK-4WT and CYK-4R459A label the membranes in the syncytial gonad, CYK-4∆C1 does not.
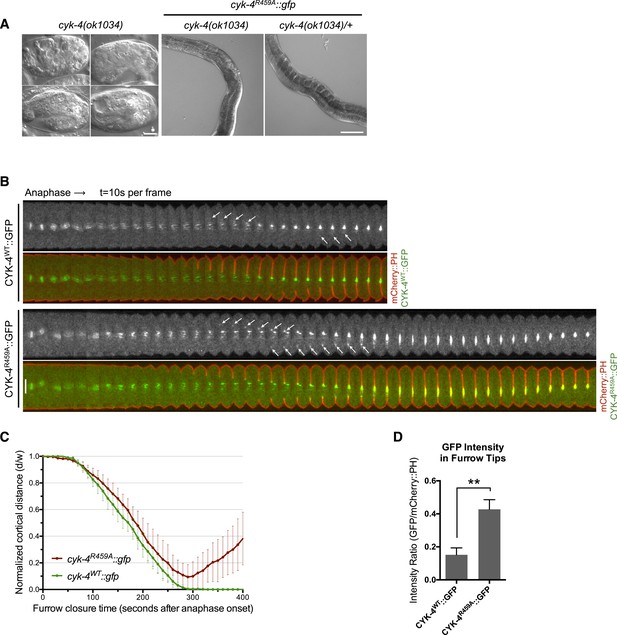
CYK-4 GAP activity is required for cytokinesis and viability.
(A) cyk-4(ok1034) null embryos arrest at variable stages during embryogenesis (left). The GAP-deficient CYK-4R459A transgene rescues the embryonic lethality, but animals arrest as sterile adults (middle). GAP-deficient CYK-4R459A is recessive, no phenotypes is seen in the presence of cyk-4(+) (right). Right scale bar 50 µm. (B) GAP-deficient CYK-4R459A embryos fail to complete cytokinesis. Kymographs are generated as in Figure 1D, with the exception that the signal from the CYK-4 transgenes is also shown (green), overlaid on mCherry::PH (red). CYK-4R459A accumulates more strongly at furrow tips as compared to CYK-4WT (arrows). (C) The kinetics of furrow ingression in CYK-4WT and CYK-4R459A embryos. Results are quantified as described in Figure 1G. (D) Membrane accumulation of CYK-4WT and CYK-4R459A. Results are quantified as described in Figure 1B (**p < 0.01, by t-test).
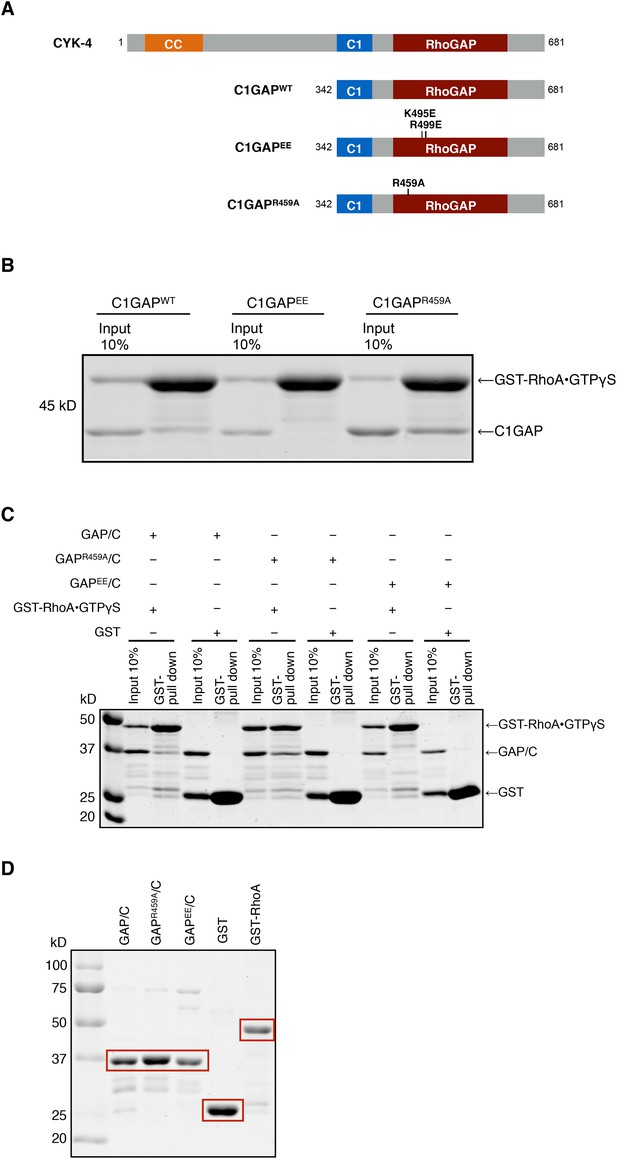
Binding of CYK-4 variants to RhoA.
(A) Schematic of the domain structure of full length CYK-4 and the recombinant GAP domains used in binding studies. The positions of the relevant mutations are indicated. (B) Binding assays between CYK-4 C1GAP domains and GST-tagged RhoA complexed with GTPγS. CYK-4WT GAP domain binds to RhoA•GTPγS. The CYK-4EE GAP domain exhibits reduced binding to RhoA•GTPγS, whereas CYK-4R459A GAP domain exhibits increased binding to RhoA•GTPγS. (C) GST-pulldown assays between GST-RhoA•GTPγS and CYK-4 GAP/C domain variants. Wild-type CYK-4 GAP binds to RhoA•GTPγS. CYK-4R459A GAP exhibits increased binding to RhoA•GTPγS, whereas CYK-4EE GAP exhibits reduced binding to RhoA•GTPγS. None of the CYK-4 GAP variants exhibit binding activity to GST alone. (D) Proteins used in binding assays in (C).

GAP activity of CYK-4 and variants.
(A) Time course of GTP hydrolysis by RhoA in the presence or absence of 200 nM CYK-4 GAP or CYK-4R459A GAP. (B) Titration of CYK-4 GAP activity towards CED-10/Rac1 and RhoA. Free phosphate was measured after 30 min.
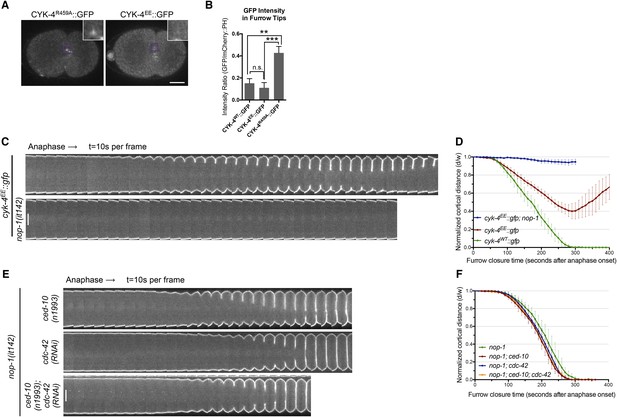
RhoA binding by CYK-4 is required for cytokinesis.
(A) CYK-4R459A, but not CYK-4EE, hyperaccumulates on the plasma membrane of the ingressing cleavage furrows (boxed regions). (B) Membrane accumulation of CYK-4::GFP variants was quantified as described in Figure 1B. (N = 8–12 embryos; **p < 0.01, by one way ANOVA followed by Tukey multiple comparison). (C) The Rho family GTPase binding defective variant of CYK-4, CYK-4EE, , causes cytokinesis defects. Kymograph analysis of the progression of cytokinesis in CYK-4EE embryos in the presence or absence of NOP-1 function. Kymographs were assembled as described in legend to Figure 1D. (D) The kinetics of furrow ingression in CYK-4EE embryos. Results are quantified as described in Figure 1G. (E) Inactivation of CDC-42 and CED-10/Rac1, either alone or in combination, does not cause cytokinesis defects in the sensitized nop-1 background. Kymographs were assembled as described in legend to Figure 1D. Note that depletion of CDC-42 results in symmetric cleavage furrow ingression. (F) The kinetics of furrow ingression in embryos deficient in nop-1 and/or CDC-42 and/or CED-10/Rac1 function. Results are quantified as described in Figure 1G.
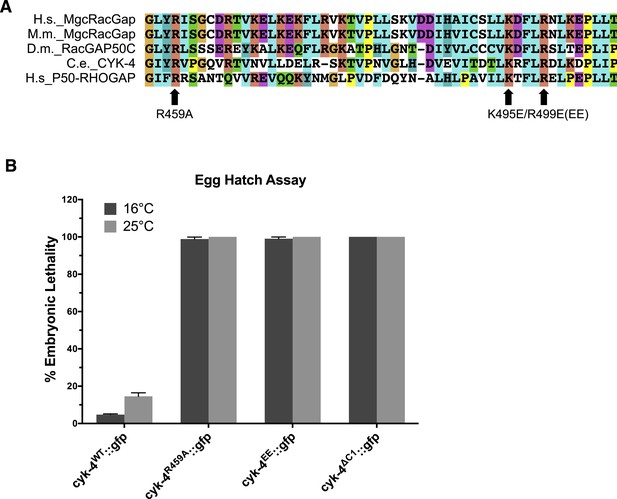
Location and conservation of mutated residues in the CYK-4 GAP domain.
(A) Alignment of a region of the GAP domain of CYK-4 from various species and the unrelated RhoGAP P50. The positions of the catalytic arginine (Ce 459) and the conserved basic residues (Ce K495/R499) are indicated. Accession numbers Q9H0H5.1; Q9WVM1.1; AAF58324.1; CAB04593.1; Q07960.1. (B) Embryonic lethality of CYK-4 variants at two temperatures. cyk-4R459A::gfp, cyk-4EE::gfp, and cyk-4∆C1::gfp exhibit fully penetrant embryonic lethality at 16°C and 25°C. cyk-4WT::gfp is included as a control.
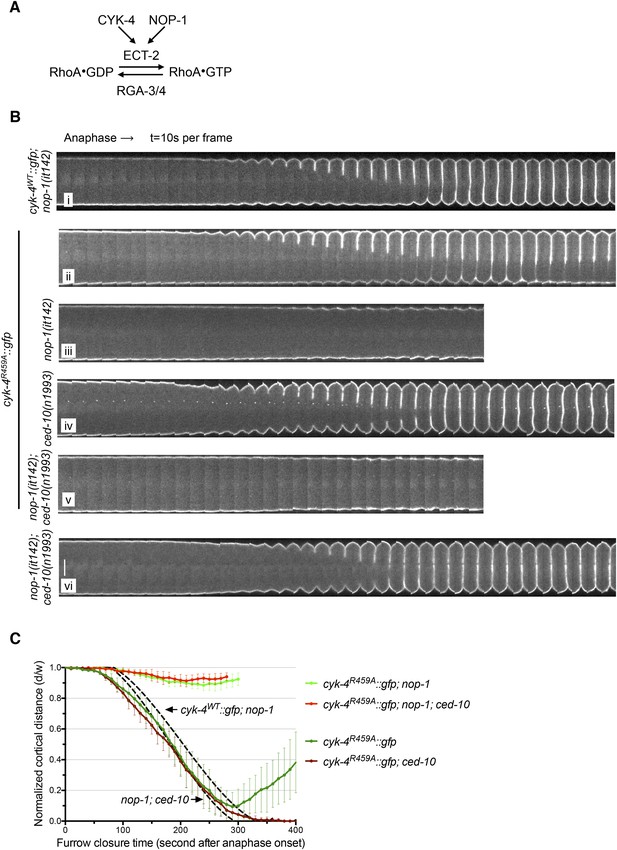
The GAP activity of CYK-4 is required for centralspindlin-dependent furrowing independent of CED-10/Rac1.
(A) Schematic depiction of the known regulators of RhoA. (B) GAP defective CYK-4 causes cytokinesis defects that are greatly enhanced by loss of NOP-1 function; this defect is not fully suppressed by mutation of ced-10/rac1. Kymograph analysis of the progression of cytokinesis in CYK-4R459A embryos in the presence or absence of CED-10/Rac1 and/or NOP-1 function. The kinetics of furrow ingression in cyk-4WT::gfp; nop-1(it142) and nop-1(it142); ced-10(n1993) embryos are shown for comparison. Kymographs were assembled as described in legend to Figure 1D. (C) The kinetics of furrow ingression in CYK-4R459A embryos. Results are quantified as described in Figure 1G. The kinetics of furrow ingression in cyk-4WT::gfp; nop-1(it142) and nop-1(it142); ced-10(n1993) embryos from Figures 1F, 3F are shown as dashed lines for comparison.
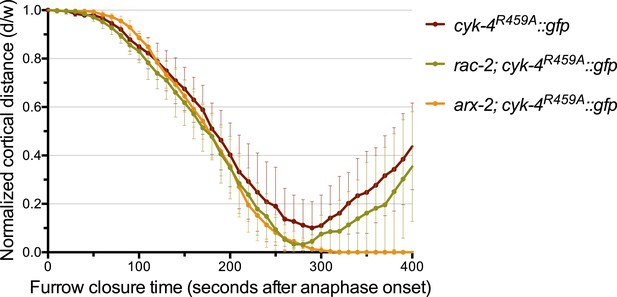
Depletion of ARX-2, but not RAC-2, suppresses the cytokinesis defect in CYK-4R459A embryos.
The kinetics of furrow ingression in CYK-4R459A embryos depleted of RAC-2 or ARX-2. Results are quantified as described in Figure 1G. The kinetics of furrow ingression in cyk-4R459A::gfp embryos from Figure 4D is shown for comparison.
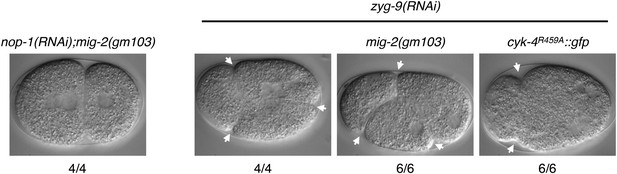
A gain of function mutation, mig-2(gm103), does not affect cytokinesis in sensitized backgrounds.
NOP-1 or ZYG-9 was depleted from mig-2(gm103) embryos and the progression of cytokinesis was followed by nomarski imaging. Cytokinesis proceeds to completion in the former case, and both anterior (left) and posterior (right) furrows are formed in the latter case. ZYG-9 was depleted from cyk-4R459A::gfp embryos for comparison demonstrating loss of the posterior furrow.
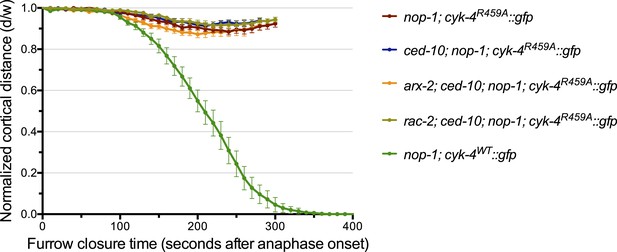
Depletion of neither ARX-2 nor RAC-2 modulates the cytokinesis defect in nop-1; ced-10; cyk-4R459A embryos.
The kinetics of furrow ingression in cyk-4R459A::gfp; nop-1(it142); ced-10(n1993) embryos depleted of either ARX-2 or RAC-2. Results are quantified as described in Figure 1G. The kinetics of furrow ingression in nop-1(it142); cyk-4WT::gfp, cyk-4R459A::gfp; nop-1(it142), and cyk-4R459A::gfp; nop-1(it142); ced-10(n1993) embryos from Figures 1G, 4C are shown for comparison.
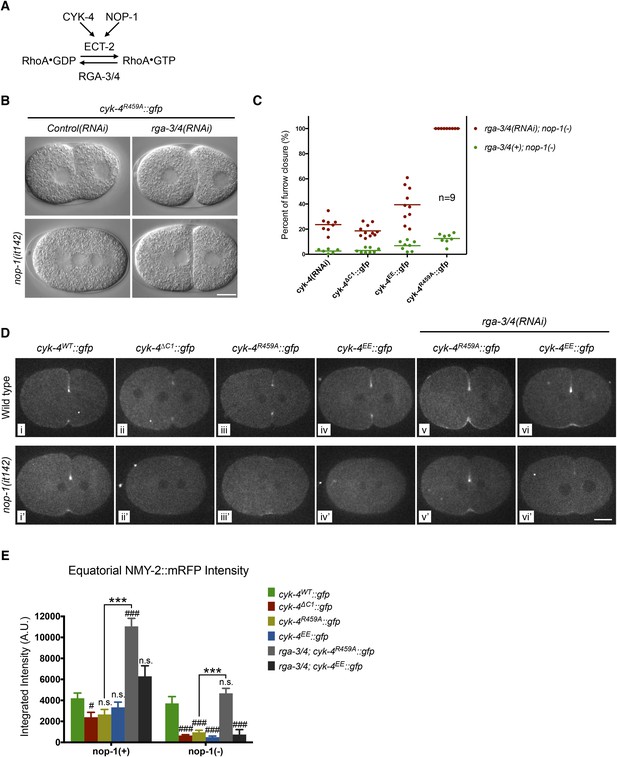
Depletion of RGA-3/4 rescues cytokinesis in CYK-4R459A embryos.
(A) Schematic depiction of the known regulators of RhoA. (B) Representative embryos demonstrating the effect of RGA-3/4 depletion on cytokinesis in CYK-4R459A embryos both in the presence and absence of NOP-1 function. (C) Depletion of RGA-3/4 rescues cytokinesis specifically in CYK-4R459A embryos. CYK-4∆C1, CYK-4EE, CYK-4R459A where expressed in nop-1(it142) embryos and the extent of furrow closure measured either in the presence (green) or the absence (red) of RGA-3/4. (D) Accumulation of the RhoA effector NMY-2 (tagged with RFP) in embryos of the indicated genotypes. Embryos are shown at ∼50% (or maximal) ingression. Note the reduction of cortical myosin accumulation in CYK-4R459A, CYK-4EE, CYK-4∆C1 embryos as compared to CYK-4WT (i–iv). The severity of this reduction is enhanced by inactivation of NOP-1 (i′–iv′). Depletion of RGA-3/4 restores myosin accumulation in CYK-4R459A and CYK-4EE embryos (v, vi), even in embryos defective in NOP-1 function (v′, vi′). (E) Quantification of total NMY-2::mRFP accumulation in the furrow region over the course of cytokinesis in embryos of the indicated genotypes. Error bars, s.e.m.; n.s. (not significant); #p < 0.05; ###p < 0.001 refers to significance relative to wild-type in nop-1(+) and nop-1(it142), respectively by one way ANOVA followed by Tukey multiple comparison. ***p < 0.001 for the indicated comparison.
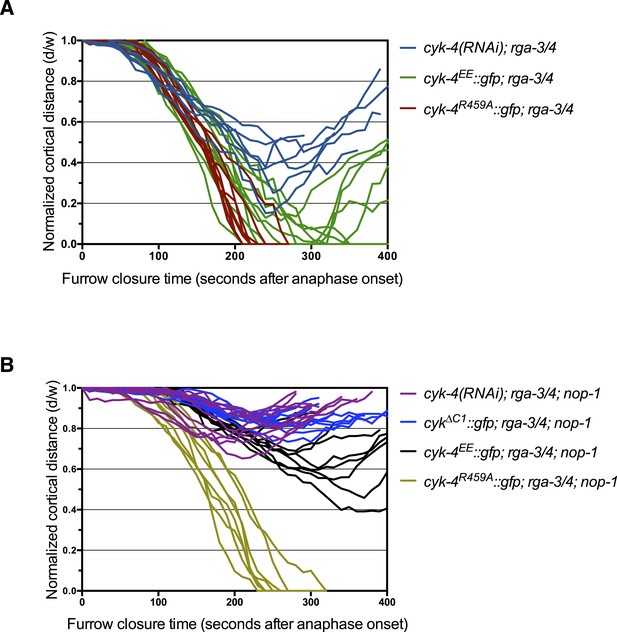
Furrow ingression of RGA-3/4-depleted embryos expressing CYK-4 variants.
(A) Line graphs depicting the kinetics of furrow ingression in (A) [cyk-4(RNAi), CYK-4∆C1, CYK-4EE, CYK-4R459A]; nop-1(it142); rga-3/4(RNAi) embryos and (B) [cyk-4(RNAi), CYK-4EE, CYK-4R459A]; rga-3/4(RNAi). These data are summarized in Figure 5C.
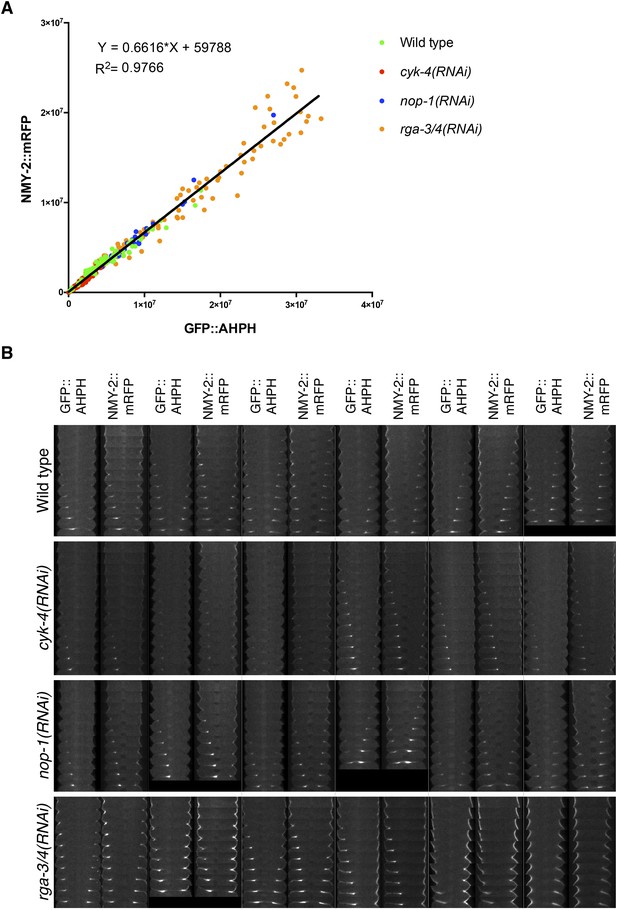
Comparison between accumulation of NMY-2::RFP and the RhoA biosensor during cytokinesis.
(A) Correlation between the accumulation of NMY-2::mRFP and GFP::AHPH (RhoA biosensor [Tse et al., 2012]) in embryos of the indicated genotypes. Each spot corresponds to the average integrated intensity of the saturation > signal >1.5*cytoplasmic background in a pair of images during cytokinesis (i.e., each embryo is represented by 9–11 dots). The data are fit to a line as shown. (B) Gallery of images showing the distribution of NMY-2::mRFP and GFP::AHPH in the furrow region during cytokinesis in sets of embryos of the indicated genotypes.
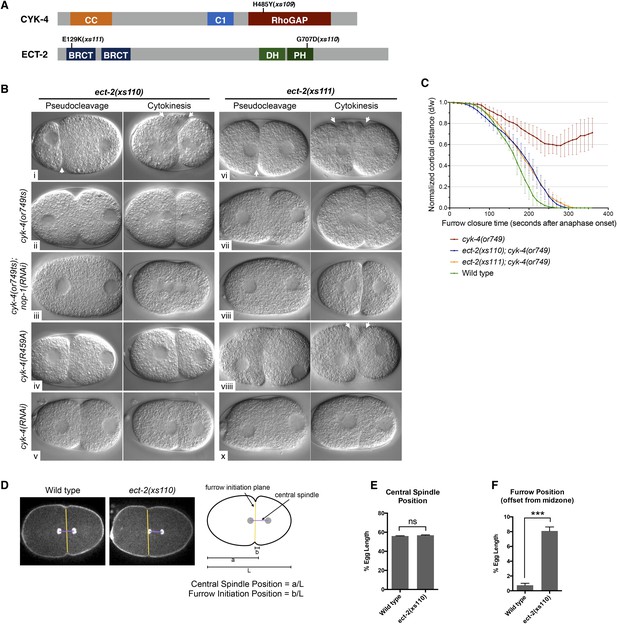
Mutations in ECT-2 suppress cyk-4(or749ts).
(A) Schematic depicting the domain structure of CYK-4 and ECT-2 with the positions of the mutations isolated in the cyk-4(or749ts) suppressor screen. (B) ect-2(xs110) and ect-2(xs111) suppresses both cyk-4(or749ts) and CYK-4R459A. Images of embryos of the indicated genotypes are shown at the pronuclear stage and during cytokinesis. Both ect-2(xs110) (i) and ect-2(xs111) (v) embryos exhibit hypercontractility (arrows) that is suppressed by cyk-4(or749ts) (ii and vii). Depletion of NOP-1 from cyk-4(or749ts); ect-2(xs110) eliminates contractility during pseudocleavage and greatly reduces contractility during cytokinesis (iii). Depletion of NOP-1 from cyk-4(or749ts); ect-2(xs111) eliminates contractility during pseudocleavage but cytokinesis is still observed (viii). ect-2(xs110) and ect-2(xs111) both allow cytokinetic completion in CYK-4R459A (iv, ix). Depletion of CYK-4 prevents completion of cytokinesis in ect-2(xs110) and ect-2(xs111) (v, x). Phenotypes shown were seen in (i) 18/18 embryos; (ii) 11/12; (iii) 7/16, 5/16 showed less contractility; (iv) 6/6; (v) 7/7; (vi) 14/14; (vii) 18/18; (viii)13/16; (ix) 6/6; (x) 6/6. (C) ect-2(xs110) and ect-2(xs111) suppress cyk-4(or749ts). The kinetics of furrow ingression in cyk-4(or749ts) embryos and in the suppressed strains. Results are quantified as described in Figure 1G. (D) ect-2(xs110) causes defects in cleavage plane positioning. The position of furrow initiation and the spindle midzone are indicated in yellow and purple, respectively (see schematic). (E) Quantification of the mean position of the central spindle (±s.e.m) as a function of egg length in wild-type and ect-2(xs110) embryos. (F) Quantification of the mean position of furrow initiation (±s.e.m) relative to the center of the spindle midzone in wild-type and ect-2(xs110) embryos. (***p < 0.001, by t-test).
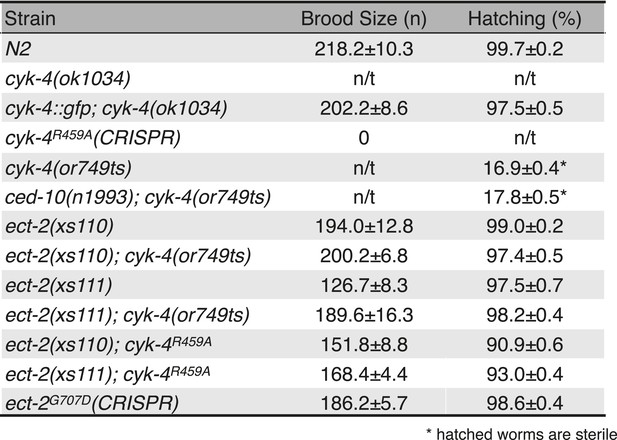
Viability and fertility of cyk-4 mutants and suppressors.
Mean brood size and hatch rates of strains of the indicated genotypes (n/t, not tested). All strains were tested at 25°C.
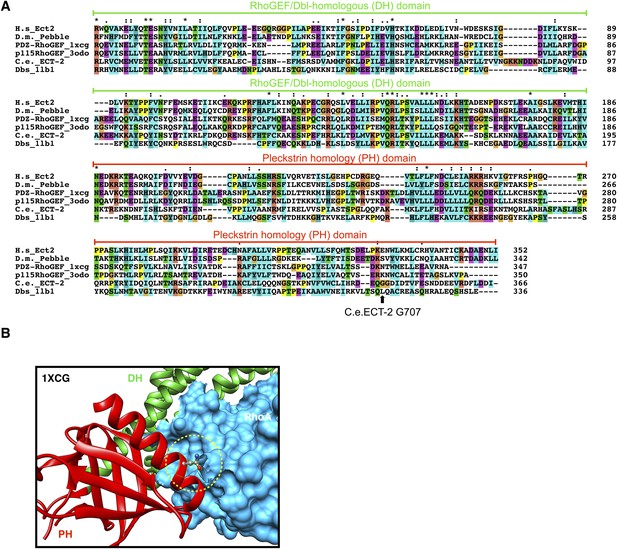
Conservation of ECT-2 GEF domain and inferred position of the ect-2(xs110) allele.
(A) Sequence alignment of DHPH domains from ECT-2 orthologs and other GEFs highlighting the position of the G707. Accession numbers: Q9H8V3.4; AAF50508.2; O15085.1; Q92888.2; CAB54311.1; Q64096.2. (B) Portion of the crystal structure (1XCG) of PDZRhoGEF (green, DH domain; red, PH domain; blue, RhoA). The equivalent to G707, N1068 is highlighted.
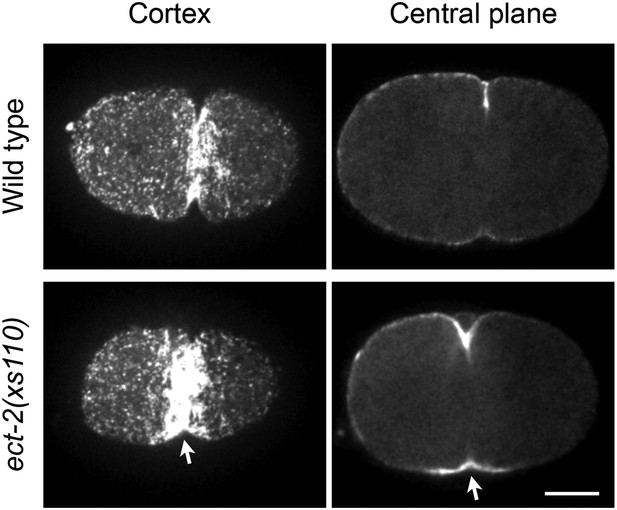
Accumulation of myosin during cytokinesis in ect-2(xs110).
ect-2(xs110) results in hyperaccumulation of NMY-2::GFP during cytokinesis.
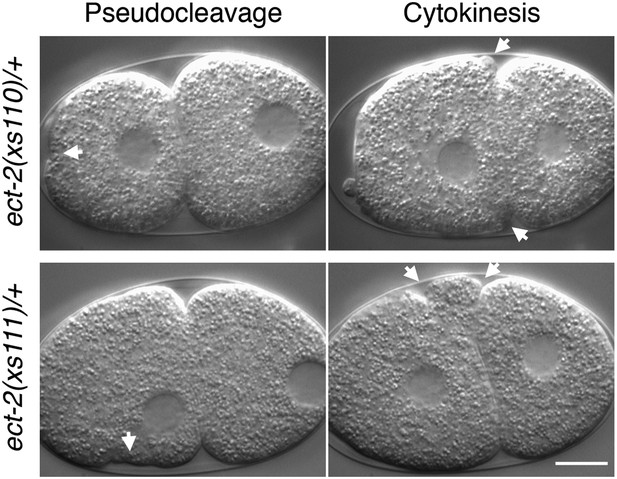
ect-2(xs110) and ect-2(xs111) are dominant gain of function mutations.
Nomarski imaging of ect-2(xs110)/+ and ect-2(xs111)/+ during pseudocleavage and cytokinesis. Hypercontractility (arrows) is observed in the presence of a wild-type allele of ect-2.
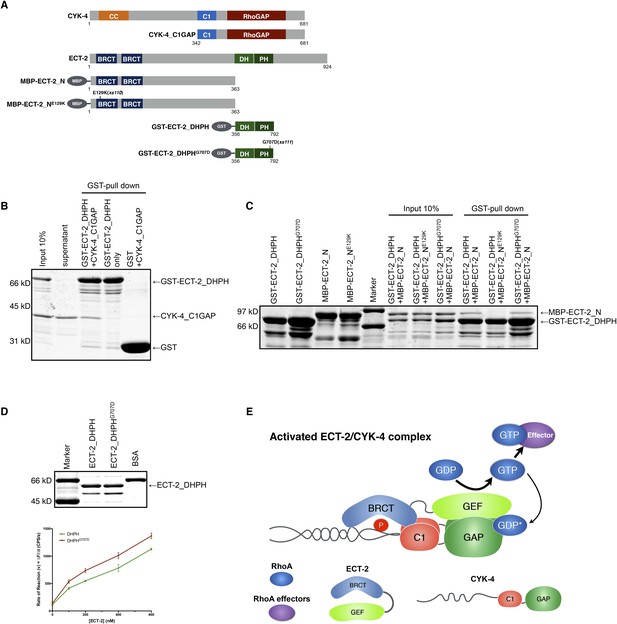
Biochemical basis for suppression by ECT-2 variants.
(A) Schematic depiction of the domain organization of CYK-4 and ECT-2 and the recombinant fragments used for biochemical analyses. (B) GST pulldown between GST-ECT-2 DHPH and CYK-4 C1GAP. (C) Pulldown assay between MBP-ECT-2-N (wild-type and the E129K variant) with GST-ECT-2 DHPH (both wild-type and the G707D variant). The MBP proteins were present in the soluble fraction and incubated with the GST-DHPH fragments bound to beads. The wild-type N-terminus associates with wild-type and G707D C-termini. However, the ECT-2 NE129K is defective in binding to wild-type C-terminus. (D) The G707D substitution activates the exchange activity of ECT-2. Exchange assays were performed with RhoA•GDP exchanging for mant-GTP at different concentrations of ECT-2 DHPH and ECT-2 DHPHG707D. Results shown are the average (±s.e.m) of three assays. The gel contains 1 µg of each ECT-2 variant and a BSA standard. (E) Working model summarizing the proposed mechanism for ECT-2 activation. Note that only the CYK-4 subunit of centralspindlin is shown. In vivo, centralspindlin is predicted to be oligomeric and the entire complex bound to the plasma membrane.
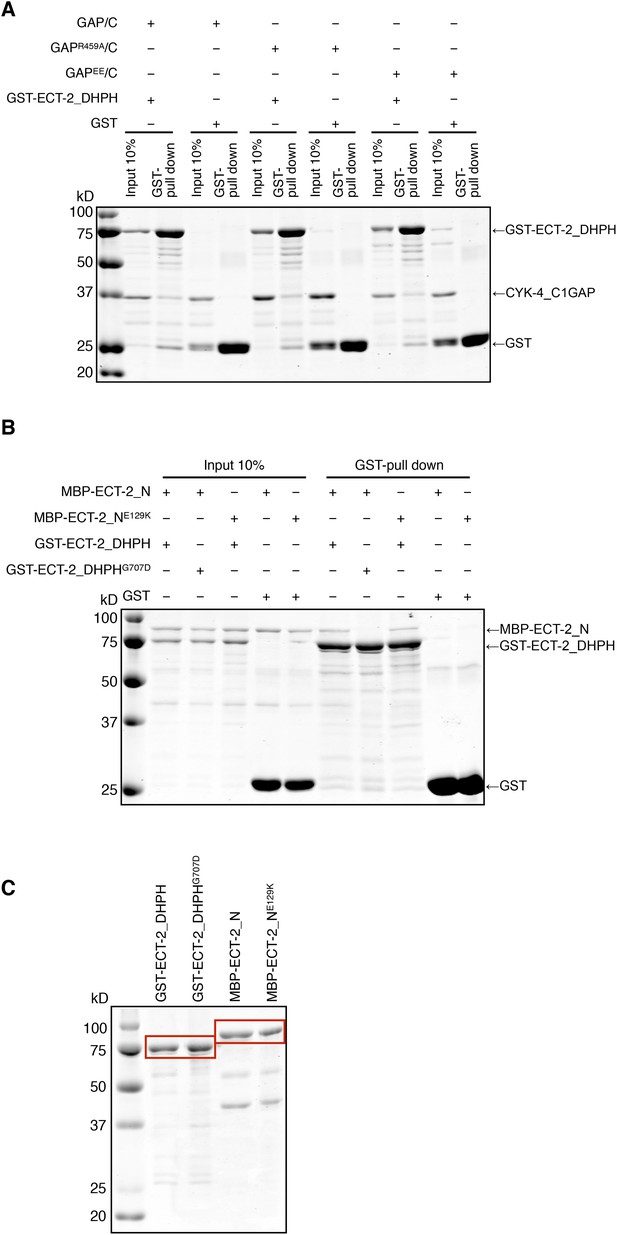
Biochemical characterization of CYK-4 and ECT-2 variants.
(A) GST-pulldown assay between GST-ECT-2 DHPH and CYK-4 GAP/C. Wild-type, R459A, and EE variants of CYK-4 GAP exhibit similar binding activity to GST-ECT-2 DHPH but do not bind to GST alone. (B) GST-pulldown assay between GST-ECT-2 DHPH (wild-type and G707D variant) and MBP- ECT-2-N (wild-type and E129K variant). The wild-type N-terminus associates with wild-type and G707D DHPH, whereas ECT-2 N E129K is defective in binding to wild-type DHPH. Neither MBP-ECT-2-N variant binds to GST alone. (C) Proteins used in binding assays.
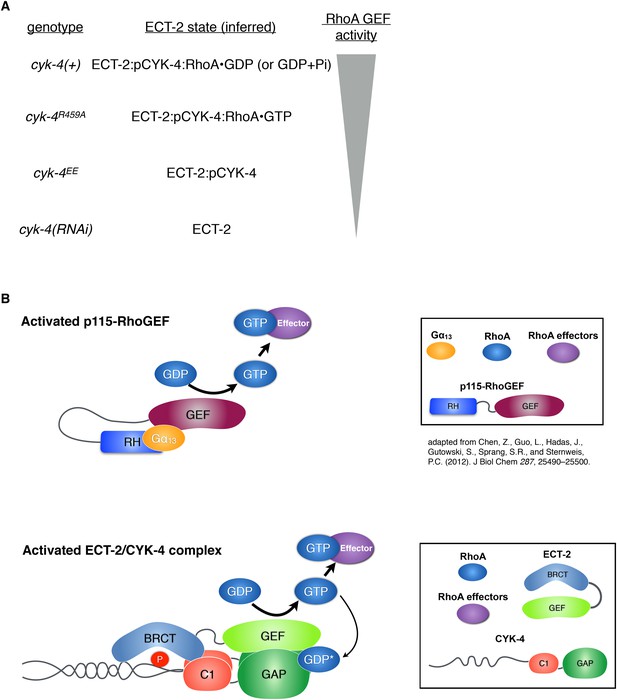
Proposed states of the ECT-2/CYK-4 complex.
(A) Correspondence between the cyk-4 genotypes, the proposed forms of the CYK-4:ECT-2 complexes, and the observed levels of contractility. (B) Schematic depicting the mechanism of activation of p115-RhoGEF by its RGS homology domain and G alpha 13 (based on small angle x-ray scattering analysis) compared to the proposed working model for ECT-2 activation.
Additional files
-
Supplementary file 1
C. elegans strains used in this study.
- https://doi.org/10.7554/eLife.08898.028
-
Supplementary file 2
List of oligonucleotides used in this study.
- https://doi.org/10.7554/eLife.08898.029





