Clusters of synaptic inputs on dendrites of layer 5 pyramidal cells in mouse visual cortex
Figures
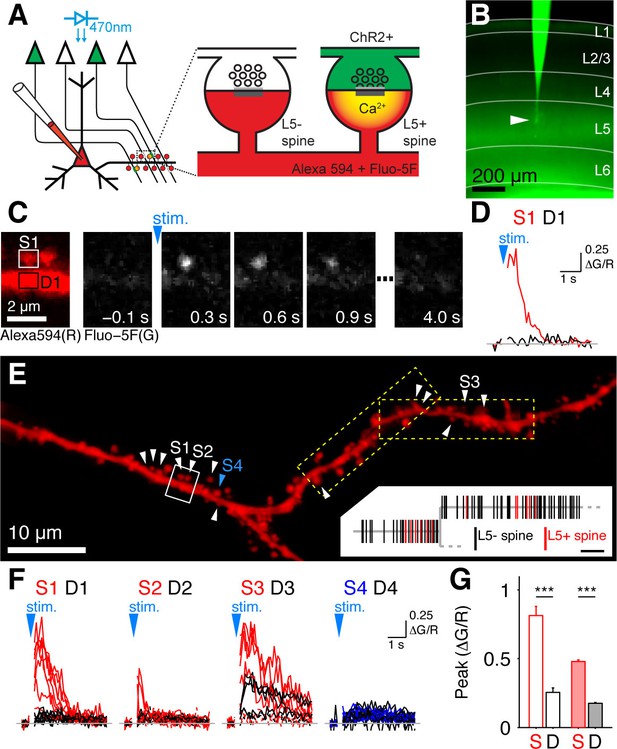
Mapping intralaminar inputs on the dendrites of L5 pyramidal neurons with channelrhodopsin-assisted synapse identity mapping (CASIM).
(A) Schematic diagram illustrating the principle of CASIM. Synapses between presynaptic L5 neurons expressing ChR2 (green) and a postsynaptic L5 neuron filled with calcium indicator (red) are identified by calcium signals in dendritic spines (L5+ spines) evoked by photostimulation of ChR2. (B) Acute slice of primary visual cortex from a Thy1-ChR2 mouse. A L5 pyramidal cell (arrowhead) was patched and filled with Fluo-5F. (C) Example of a spine calcium signal evoked by ChR2 activation: Left, image of two spines and their parent dendrite (from white box in panel E) in the red channel (Alexa 594) for structural imaging. Right, sequence of frames from calcium imaging in one trial showing calcium signal in the left spine. (D) Ca2+ signal corresponding to panel C (ΔG/R; G, Fluo-5F; R, Alexa 594; red, spine; black, dendrite at spine base). (E) Analyzed dendritic stretch from cell in B. L5+ spines are marked by white arrowheads. Blue arrow head marks an example L5- spine. Dashed boxes indicate the size of imaging tiles. Inset, corresponding dendrogram; scale bar, 10 µm. (F) Ca2+ signals from all trials recorded in a selection of L5+ spines (S1 to S3), one L5- spine (S4) and at their respective dendritic bases (D1-D4) in E. (G) Peak ΔG/R of L5+ spines (S) and dendrites (D) in cell in B (blank bars; n = 13 spines) and in all cells (filled bars; n = 199 spines).
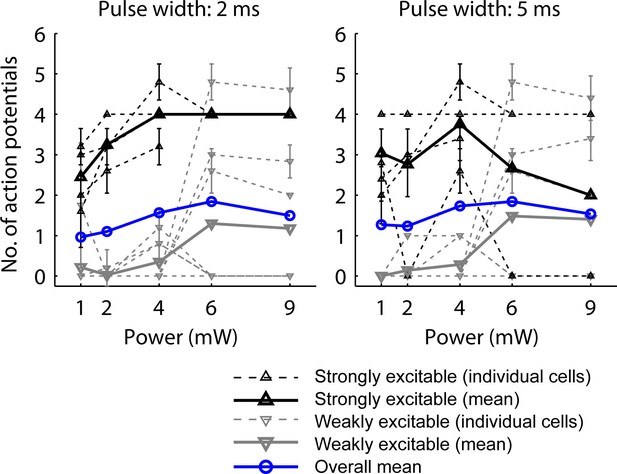
Characterization and calibration of optical stimulation.
The photostimulus protocol was optimized to maximize the probability of synaptic transmission. 3 pulses at 30 Hz were applied to deliver the photostimulus in the duration of one frame (100 ms) while the PMTs for imaging were protected with a shutter. Calibration curves were acquired at 2 (left) and 5 ms (right) pulse width on L5 pyramidal neurons selected blindly (n = 12). Number of APs was measured in cell-attached mode. The mean overall calibration curves (blue) did not show a uniform increase with photostimulation power, but a declining AP yield at high powers. To analyze whether this may be explained by potential adverse effects of strong photostimulation on strongly driven neurons, we analyzed the calibration curves with respect to two populations of neurons: Strongly excitable neurons (with more than one AP on average at 1, 2 and 4 mW power; black; dashed, individual cells; solid, mean curve; n = 4 cells at 2 ms pulse width, n = 5 cells at 5 ms pulse width) showed saturation above 4 mW power with 2 ms pulse width and reduced activity at high illumination intensity at 5 ms pulse width. Weakly excitable neurons (gray; dashed, individual cells; solid, mean curve; n = 8 cells at 2 ms pulse width, n = 7 cells at 5 ms pulse width) were not affected adversely by illumination. Therefore for mapping, we chose 2 ms pulses at 4 mW as stimulus, which activates but does not overexcite weakly and strongly excitable neurons, respectively. Error bars correspond to standard deviation.
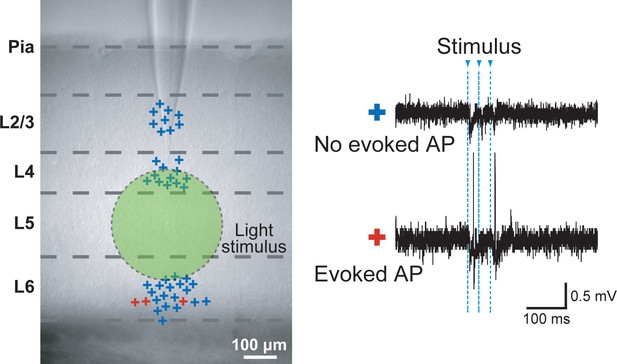
Specificity of evoked presynaptic activity.
AP generation in neurons outside of L5 was measured by cell attached recordings obtained during a wide-field light stimulus in L5 (green circle). Left panel: marks (+) indicate the vertical location of the cells which were recorded in slices from Thy1-ChR2 mice. Color codes for the neurons with no evoked APs (blue) and evoked APs (red). Right panel: example traces. In total 45 cells were recorded, of those only 3 cells in L6 exhibited light evoked activity (6.7%).
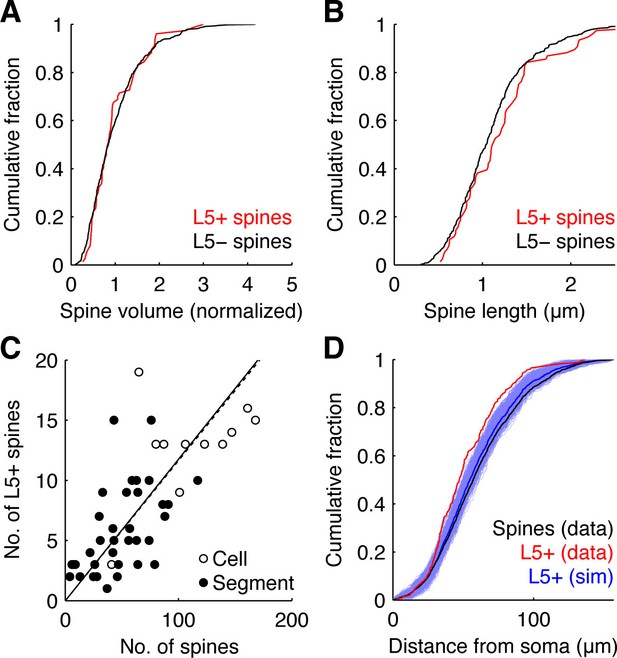
Characterization of L5+ spines.
(A,B) Cumulative fraction of spine volume (A) and normalized spine length (B) of L5+ (red) and L5- spines (black). Spine volume and length were measured only from spines extending laterally in image stacks, since spines aligned along z could not reliably be separated from the dendrite. For spine volume, the summed intensity of pixels enclosed by the respective ROI was calculated for each z-plane after background subtraction. The highest sum represented the volume of the spine. Measured volumes were normalized to the mean spine volume within dendritic segments between branching points. Spine lengths were measured on maximum z-projections as the distance between the tip of the spine head and where it attached on the dendrite. (C) Number of L5+ spines versus total number of spines probed per cell (open circles; correlation, dotted line, slope = 0.12, R = 0.93) and per segment (filled circles; correlation, continuous line, slope = 0.12, R = 0.89). (D) Distribution of L5+ spine distances from the soma (red) in the experiments and from a simulation with 1000 randomly reshuffled distributions of the same number of inputs over all recorded spine positions (blue, individual simulations; dark blue, mean). For comparison also the distribution of distances of all spines from the soma is shown (black).
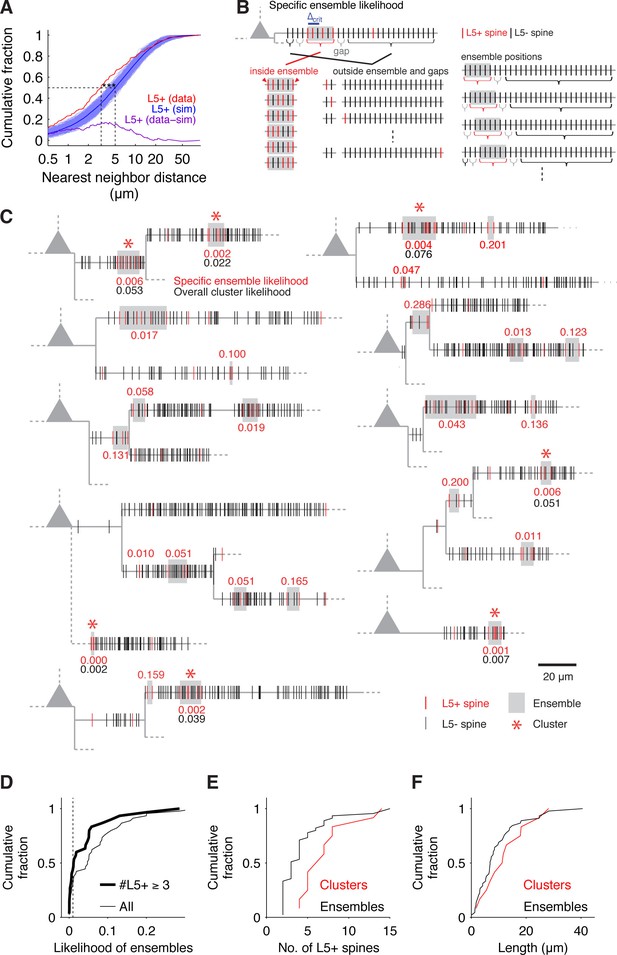
Spatial input organization.
(A) Distribution of nearest neighbor distances between L5+ spines from experiments (red) and from 1000 times random reshuffling the recorded number of inputs over all present spines within each individual dendritic segment (blue, dark blue mean trace). Median values of recorded and reshuffled data are significantly different (data, median = 3.10 μm; simulations, median = 5.02 ± 0.43 μm; p<0.0001).The plot of the difference between inter-input distances from experiments and simulations (purple) shows that the deviation is most pronounced for small inter-input distances below 10 µm with a peak deviation around 5 µm, suggesting a clustered arrangement of L5+ spines. (B) Combinatorial cluster analysis: Illustration of the principle of calculating the specific ensemble likelihood (combinatorial cluster analysis step 2). Top, example of a dendritic segment with N = 30 spines in total and n = 5 L5+ spines (red). m = 4 L5+ spines are part of an input ensemble of size M = 6 spines (gray box) by fulfilling the maximal nearest neighbor distance criterion of △crit = 2 (blue bar; cluster analysis step 1). The ensemble is flanked by 'gaps' of L5- spines (gray brackets). As illustrated below, the likelihood for observing an ensemble of M = 6 spines with m = 4 L5+ spines is determined by three factors: The number or ways the L5+ spines can be distributed inside the ensemble, the number of ways the remaining L5+ spines can be distributed outside of the ensemble and the number of ways an ensemble of the given size can be placed on the dendritic segment. Bottom left, all possible arrangements of m = 4 L5+ spines in an ensemble of M = 6 spines, note that the ensemble edges (red arrowheads) have to be occupied always to delimit the ensemble. Bottom middle, four examples of the possible assignments of the remaining n-m = 1 L5+ spine to the spines outside of ensemble and gaps. Bottom right, four examples of the possibilities to place the ensemble of size M = 6 spines into the dendritic segment with N = 30 spines in total. Note that at positions close to the beginning and end of the segment, the leading or trailing gap, respectively, are not fully realized. (C) Dendrograms of 10 out of 20 mapped pyramidal cells summarizing 7 observed input clusters (L5+ spines, red; L5- spines, black; dendrograms of all other recorded cells with input clusters in Figure 2—figure supplement 1). Gray boxes mark input ensembles (cluster analysis step 1), red numbers are the specific ensemble likelihood values (cluster analysis step 2), red asterisks mark ensembles identified as clusters (specific ensemble likelihood ≤0.01, inputs ≥3; cluster analysis step 3), black numbers are overall cluster likelihood values for each cluster (cluster analysis step 4). Note that spines, which are located very close to each other, are not resolved at this scale and their marks appear to overlap; dashed lines indicate unmapped stretches). (D) Specific ensemble likelihood scores of all ensembles (thin line) and those with at least three inputs (thick line). Dashed line, cluster likelihood threshold. (E) Number of L5+ spines in individual clusters (red) and all ensembles (black). (F) Length of clusters (red) and all ensembles (black).
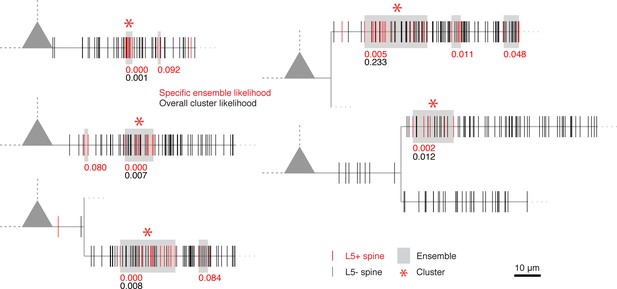
Dendrograms of all other recorded cells with input clusters not shown in Figure 2.
https://doi.org/10.7554/eLife.09222.008
Combinatorial cluster analysis: Example of ensemble types and translation to non-uniform inter spine distances.
(A) Combinatorial cluster analysis step 4: Example of all ensemble types possible with a total of n = 5 inputs (L5+ spines) and a gap/nearest neighbor distance criterion of △crit = 2 to be considered for calculating the overall cluster likelihood. (B) Translation of ensemble definition from uniform to non-uniform inter spine distances: Example of a dendritic segment with irregularly spaced spines. 5 spines receive the specific input (red, L5+), 4 of which are part of an ensemble (gray square) by complying with the nearest neighbor distance criterion △crit (indicated by the blue bar). The length of the leading and trailing edge, which flank the ensemble, corresponds to the nearest neighbor distance criterion. No spine located within these gaps receives the specific input.
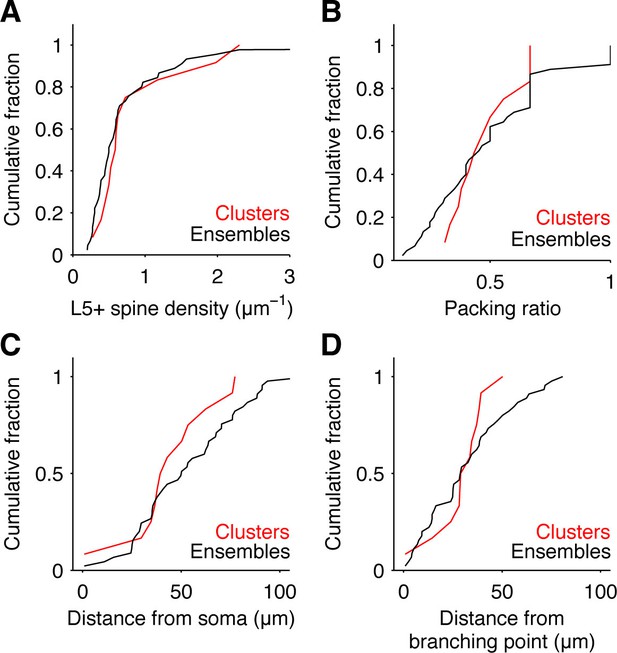
Additional properties of input clusters.
Cumulative fraction of input density (A), input packing ratio, i.e. ratio of L5+ spines over total number of spines in cluster or ensemble (B), and location as distance from soma (C) or distance from proximal branching point (D) of detected clusters (red) and all ensembles (black).
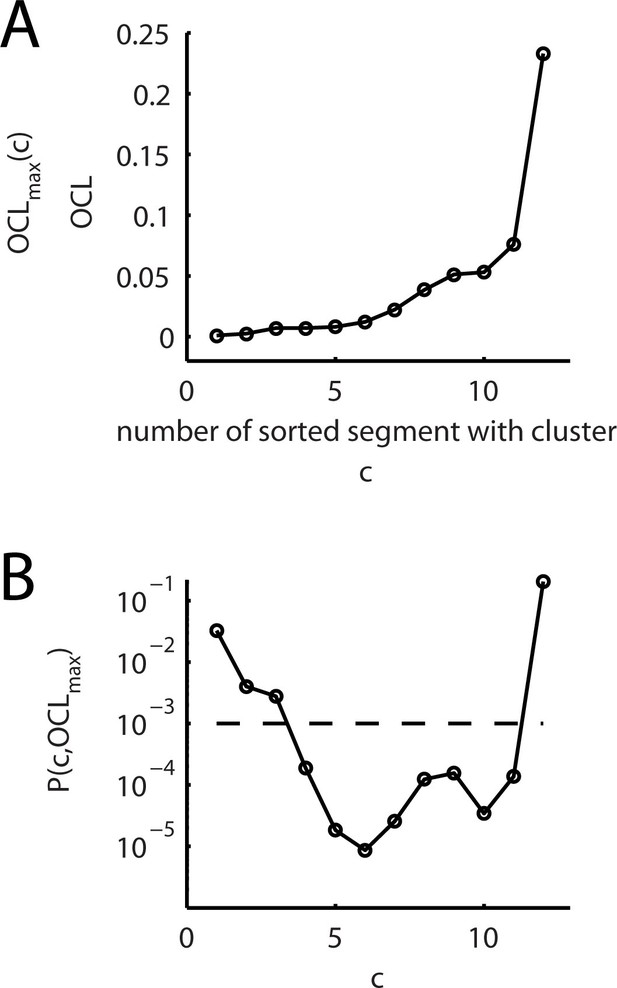
Comparison of the observed and expected numbers of dendritic segments containing an input cluster (cluster analysis step 5).
(A) For dendritic segments, which contain a synapse cluster, the overall cluster likelihood OCL is plotted against the segment number sorted by increasing OCL value. For sets of segments with OCL values below an upper bound, this graph is the same as plotting the upper bound OCLmax(c) as function of the number c of segments with OCL values below this upper bound. (B) P value P(c,OCLmax) (Equation 4) for the binomial test of the null hypothesis that the number of observed segments with an input cluster arises from a random input distribution plotted against the recorded number of segments c containing a synapse cluster. The broken line indicates the significance level of p=0.001.
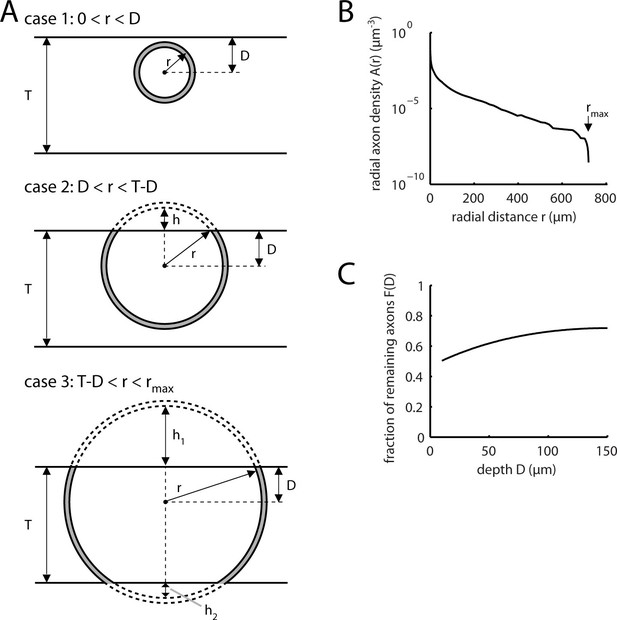
Estimation of missed L5 inputs due to slicing.
(A) Diagrams illustrating three cases to be considered for calculating the axon density at a specific dendritic site at depth D in a slice of thickness T arising from neurons at different radial distances r from this site: Case 1, shell volume completely contained in slice (top panel, see Equation 6); case 2, top cap of shell missing from slice (middle panel, see Equation 7.); case 3, top and bottom caps of shell missing from slice (bottom panel, see equ. 8). (B) Radial axon density A(r) of L5 pyramidal cell axons in mouse visual cortex based on the sholl analysis by Blackman et al., 2014 (Figure 2C, 2nd panel from left) calculated according to Equation 5. (C) Remaining fraction of axonal density within a slice of thickness T as a function of the depth D in the slice calculated according to Equation 9.
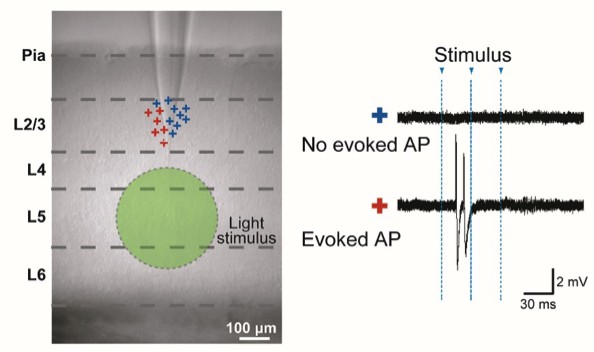
Analogous to data in Figure 1—figure supplement 2: “Specificity of Evoked Presynaptic Activity” but here without application of NBQX.
AP generation in neurons in L2/3 measured by cell attached recordings during a wide-field light stimulus in L5 (green circle). Left panel: marks (+) indicate the vertical location of the cells. Color codes for the neurons with no evoked APs (blue) and evoked APs (red). Right panel: example traces.
Additional files
-
Source code 1
Custom software in Matlab for data analysis.
- https://doi.org/10.7554/eLife.09222.013





