A robust transcriptional program in newts undergoing multiple events of lens regeneration throughout their lifespan
Figures
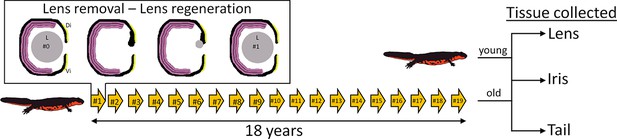
Experimental overview.
Arrows depict the number of repeated lentectomies performed over a period of 18 years. Panel shows the process of lens regeneration that occurred after each lens removal highlighted as a single arrow. At the end of the experiment, lens, iris, and tail tissues were collected from both old newts that had regenerated their lenses 19 times and young newts that had never experienced lentectomy. Di: Dorsal iris; Vi: Ventral iris; L: Lens.
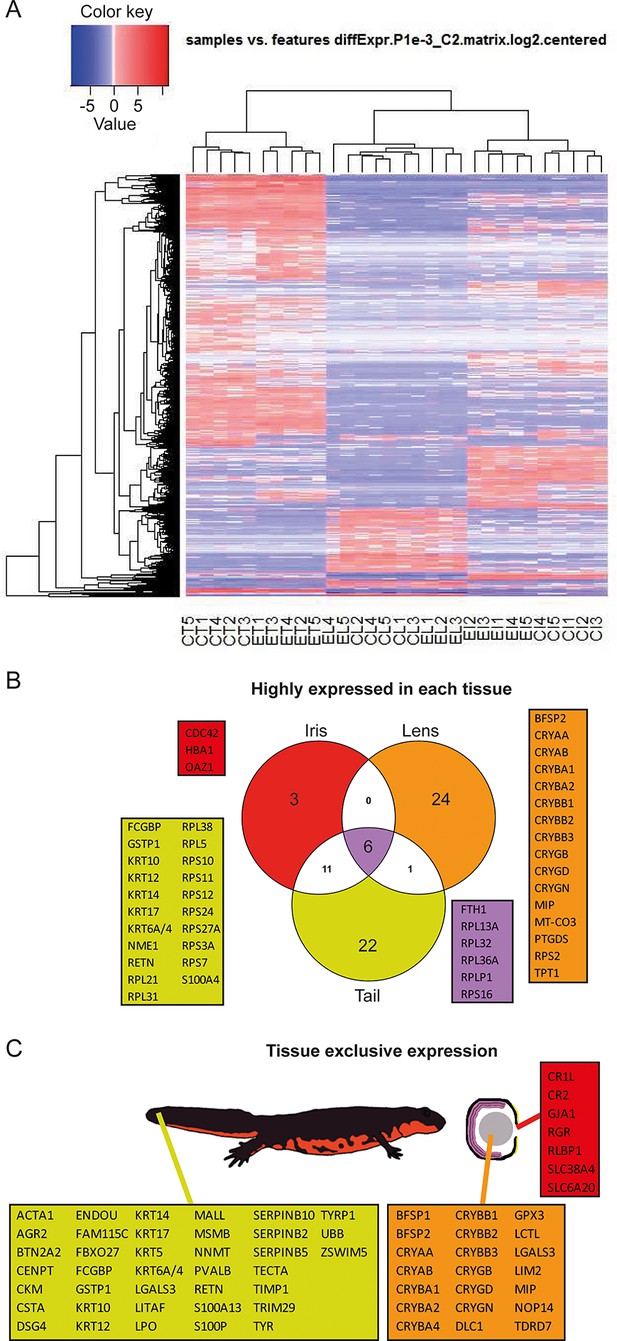
Gene expression among tissue samples.
(A) Heat map constructed from the expression profiles of the 30 sequenced samples. CT, CL, CI: #0 (control) tail, lens, and iris, respectively. ET, EL, EI: #19 (experimental) tail, lens, and iris, respectively. Genes selected based on the following parameters: p-value<=0.001 and log2(FC)>=2. Note the nearly uniform pattern between the #0 and #19 lens samples which indicates no differences between non-regenerated young lenses and repeatedly regenerated lenses from aged newts. (B) Highly expressed genes in each tissue type irrespective of age. Red, orange, and yellow colors denote genes from iris, lens, and tail samples, respectively. Comparisons are visualized using a venn graph while non-redundant annotations are highlighted using boxes including the corresponding gene names. Purple color is used for highly expressed genes in all three samples. (C) Genes that are preferentially expressed in a given tissue versus the others are denoted using the same color code as in B. The different tissues are indicated using a cartoon newt and an enlarged cross-sectioned eye.
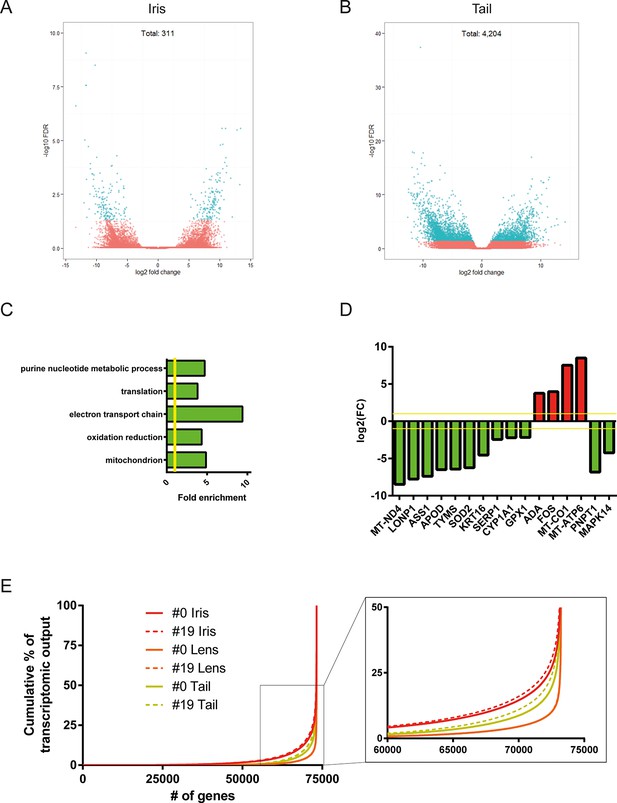
Differential gene expression between #19 and #0 tissues.
(A) Volcano plot for the #19 versus #0 iris samples. (B) Volcano plot for the #19 versus #0 tail samples. Differentially expressed genes (false discovery rate [FDR]<0.05 and fold change [FC]>2) are depicted in cyan. Tail samples, which never experienced regeneration, showed the most differentially expressed genes. Iris samples, which as the source of lens regeneration have experienced some degree of regeneration/replenishment, showed a reduced number of differentially expressed genes and an intermediate ageing profile. (C) Selected enriched (FDR <0.05) Gene Onthology (GO) terms in tail samples plotted based on their fold enrichment. Green-colored bars mark gene groups that are down-regulated in the #19 samples. Yellow line marks fold enrichment of 1. Electron transport chain is one of the functional group enriched in the down-regulated group, a well-documented ageing signature in other vertebrates. (D) Genes selected for their role in ageing and/or senescence and plotted based on their fold change between #19 and #0 tail samples. Green and red bars mark down-regulated or up-regulated genes in #19 samples, respectively. Yellow line marks log2(FC) of 1. These data provide additional evidence of ageing signs in our #19 tails samples. (E) Transcriptomic complexity between #0 and #19 tissues. Red, orange, and yellow denotes iris, lens, and tail samples, respectively. Solid and dotted lines represent tissues from #0 and #19 newts, respectively. In this graph, genes were sorted based on their expression and plotted based on their cumulative percent contribution to the overall transcriptomal output. Iris had the most complex transcriptome by having more genes contributing to the overall output (steeper line), followed by the tail and lens. Tissues from #19 newts showed slightly increased transcriptomic complexity versus their #0 counterparts (enlarged insert). Lens showed the least increase in complexity which further supports that lens regeneration is a robust process that can faithfully proceed throughout life.
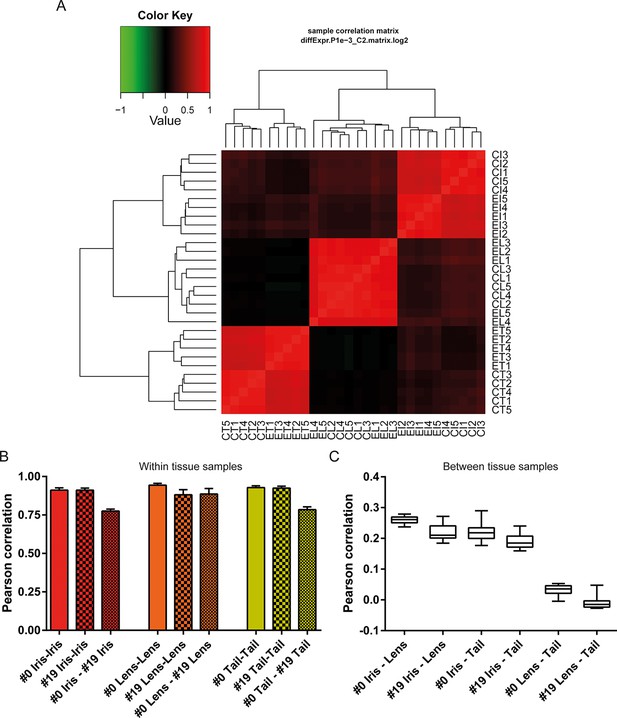
Sample correlations between the 30 samples.
(A) Sample correlation matrix plot. Note the uniform red color between #19 (experimental lens, EL) and #0 (control lens, CL), indicating high correlation between them. Iris and tail #0 and #19 samples segregate clearly creating a characteristic four-box pattern in the two edges of the plot. #19 and #0 lens samples are so similar that the cladogram clusters them together. EL4 sample exhibits the least correlation among the #19 newt lenses. (B) Within tissue correlation plotted as bar graphs for better visualization. Solid colored bars (inter-#0 correlations) and big-dotted bars (inter-#19 correlations) showed very high values. Intra #0 - #19 correlation values were lower except those in lens samples. These data indicate that #0 and #19 lens samples are very similar in regards to gene expression versus equivalent comparisons in iris and tail samples. (C) Correlations between tissues illustrated as box plots. Iris-lens gene expression correlations were the highest followed by iris-tail and lens-tail.
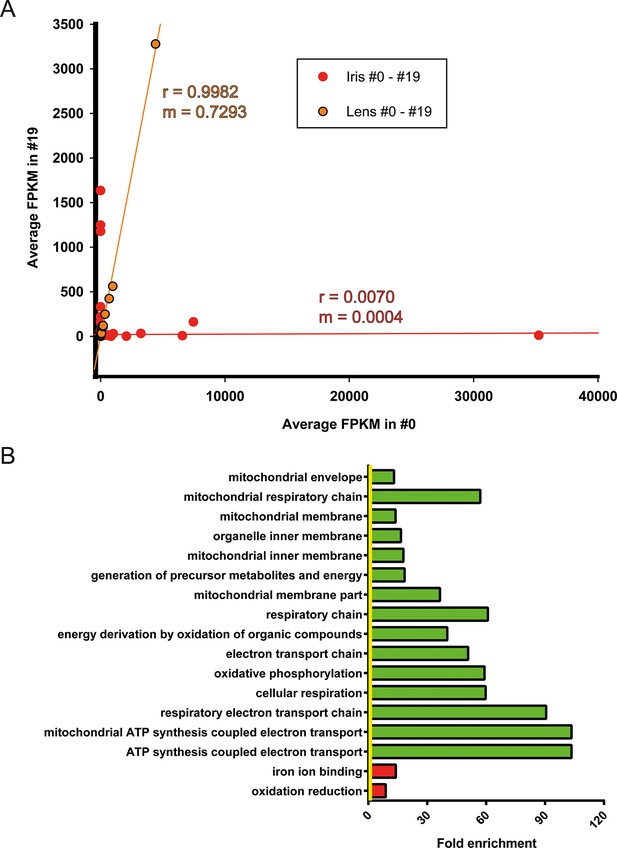
Correction of iris-regulated gene expression in the regenerated lens.
(A) The 311 genes used for this analysis were differentially regulated in #0 versus #19 iris samples. Since lens is regenerated from the dorsal iris, the question arises whether or not these differences in gene expression are reflected in the regenerated lenses. Average fragment per kilobase per millions of reads (FPKM) values of these genes from iris and lens samples were plotted in the same graph. Red and orange colors mark the iris and lens, respectively. Linear regression analysis revealed that lens genes are more correlated (r = 0.9982) with m = 0.7293 while iris genes are not correlated (r = 0.0070) as expected. Based on the slope (m) values (where m = 1 is the absolute perfect when #0 and #19 values are identical), these data indicate that the #0 and #19 lens FPKM values differ approximately 27% while the equivalent iris samples are completely different. (B) Gene Ontology enrichment analysis of genes differentially expressed between #0 and #19 iris samples. Green and red bars denote gene groups in the down-regulated and up-regulated datasets, respectively. Yellow line marks fold enrichment of 1. Note that as with the tail samples, electron transport chain is also down-regulated in these samples, a sign of ageing.

Lenses from #0 control (left) and #19 experimental (right) newts.
Note that the size, fiber arrangement, and transparency of both samples are normal.
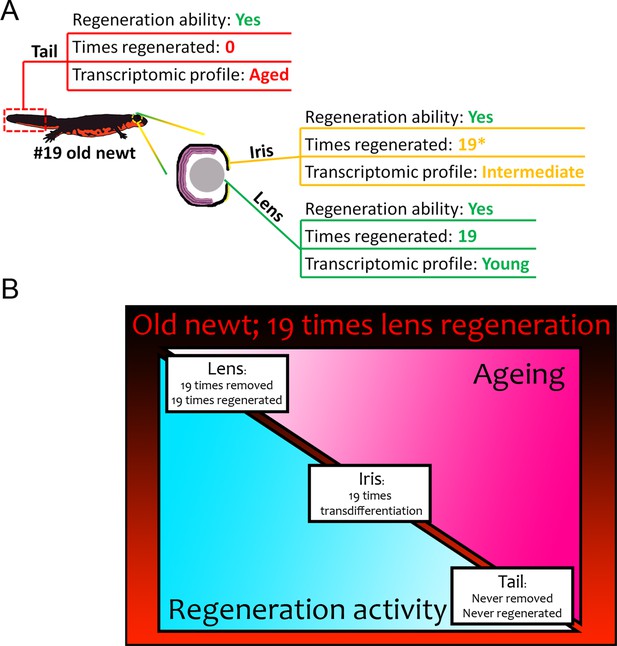
Summary of results from our transcriptomic comparisons between #19 and #0 newts.
(A) Tail samples that had never experienced regeneration showed a marked deregulation of electron transport chain, mitochondrion, and ribosome genes, in #19 newts all signatures of ageing. On the contrary, lenses that were regenerated 19 times over a period of 18 years, showed a transcriptomic profile comparable to never-regenerated lenses from young newts. Iris showed an intermediate profile marked by deregulation of electron transport chain-related genes. (B) Regeneration versus ageing in newts. Triangles indicate the amount of regeneration activity (in light blue, decreasing from left to right) and ageing signatures as found by our transcriptomic analysis (in hot pink, increasing from left to right) of sampled #19 tissues. Regeneration initiates a robust transcriptomic program that can be faithfully restarted during repeated insult with no transcriptomic deregulation or molecular signatures of ageing. In our #19 newts, lenses had been fully removed and regenerated 19 times, thus having the highest regeneration activity and showed no signs of ageing. Iris, as the source of lens regeneration, has been regenerated/replenished after transdifferentiation to lens, thus showing some activity and an intermediate profile (the asterisk indicates that iris is not regenerated completely). Tails were never removed or regenerated and showed the most deregulated genes and signatures of ageing compared to the young controls.
Additional files
-
Supplementary file 1
FPKM values of all annotated transcripts and analysis of tissue expression.
- https://doi.org/10.7554/eLife.09594.010
-
Supplementary file 2
Differential gene expression between lens tissue from repeatedly regenerated lens samples from aged newts and young newts.
- https://doi.org/10.7554/eLife.09594.011
-
Supplementary file 3
Differential gene expression between aged and young iris samples.
- https://doi.org/10.7554/eLife.09594.012
-
Supplementary file 4
Differential gene expression between aged and young tail samples.
- https://doi.org/10.7554/eLife.09594.013
-
Supplementary file 5
Annotation and gene ontology analysis of differentially expressed genes in the tail samples.
- https://doi.org/10.7554/eLife.09594.014
-
Supplementary file 6
Correlation values between all samples.
- https://doi.org/10.7554/eLife.09594.015
-
Supplementary file 7
Correlation plot using jackknifing method.
- https://doi.org/10.7554/eLife.09594.016
-
Supplementary file 8
Correlation plot using 20% random sampling method.
- https://doi.org/10.7554/eLife.09594.017
-
Supplementary file 9
Analysis of EL4 gene expression to determine signs of ageing.
- https://doi.org/10.7554/eLife.09594.018
-
Supplementary file 10
Potential correction of the iris profile upon transdifferentiation to lens.
- https://doi.org/10.7554/eLife.09594.019
-
Supplementary file 11
Annotation and gene ontology analysis of differentially expressed genes in the iris samples.
- https://doi.org/10.7554/eLife.09594.020





