A network of autism linked genes stabilizes two pools of synaptic GABAA receptors
Figures
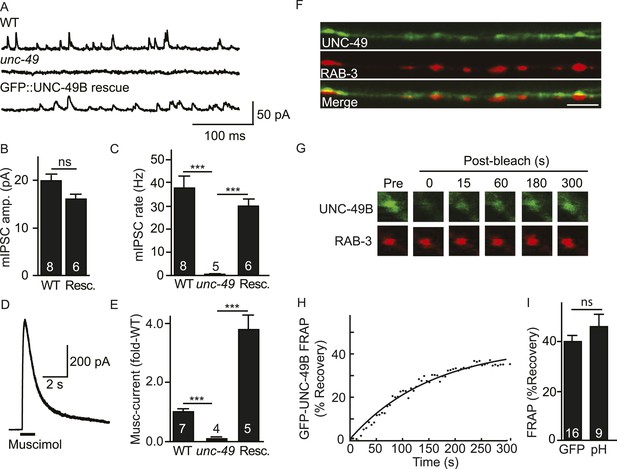
Inhibitory NMJs contain both mobile and immobile UNC-49/GABAA receptors.
(A–E) mIPSCs and muscimol-evoked currents were abolished in unc-49 mutants, and were restored by a transgene expressing GFP-tagged UNC-49B in body muscles. mIPSCs (A–C) and muscimol-evoked currents (D, E) were recorded from adult body wall muscles. For mIPSCs, representative traces (A), mean current amplitude (B) and mean frequency (C) are shown. For muscimol-evoked currents, a representative wild type response (D), and mean current amplitude (E) are shown. GFP-tagged UNC-49B is localized to GABAergic NMJs. (F) The distribution of muscle expressed GFP-UNC-49B (Green) is compared to presynaptic RAB-3::mCherry (Red), expressed in GABAergic motor neurons (scale bar 5 μm). (G–I) Synaptic UNC-49B consists of both mobile and immobilized receptors. The mobility of synaptic GFP-UNC-49B and pHluorin-tagged UNC-49B (pH-UNC-49B) was analyzed by FRAP. Representative images of GFP-UNC-49B FRAP (G), a representative scatter plot of GFP-UNC-49B fluorescence recovery (solid line indicates a single exponential fit) (H), and summary data for fluorescence recovery of GFP- and pH-UNC-49B (I) are shown. Examples of scatter plots for pH-UNC-49B recovery are shown in Figure 4—figure supplement 1A. Pre-synaptic RAB-3::mCherry fluorescence was captured as control. Values that differ significantly are indicated (***, p < 0.001; ns, not significant). The number of animals analysed is indicated for each genotype. Error bars indicate SEM.
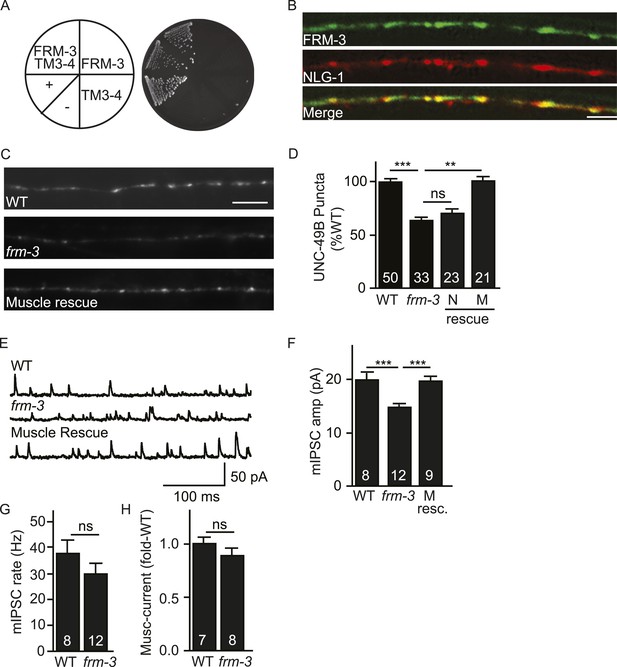
FRM-3 EPB4.1 binds UNC-49B and is required for its synaptic targeting.
(A) FRM-3′s ERM domain binds the UNC-49B TM3-4 loop in yeast 2-hybrid assays. Growth of Y2HGold cells on selective media (–Trp/-Leu/-His/-Ade) is shown. Yeast cells were transformed with vectors expressing the indicated fusion proteins. Positive (+, pGBKT7-53 and pGADT7-T) and negative (−,pGBKT7-Lam and pGADT7-T) controls are indicated. (B) Muscle expressed FRM-3::GFP (Green) and NLG-1::mCherry (Red) are co-localized in the nerve cord (scale bar 5 μm). (C, D) GFP-UNC-49B puncta fluorescence in the nerve cord was decreased in frm-3 mutants. This defect was rescued by transgenes expressing FRM-3 in body muscles (M) but not by those expressed in GABAergic neurons (N). Representative images (C, scale bar 5 μm) and mean puncta intensity (D) are shown. (E–G) mIPSC amplitude was decreased in frm-3 mutants and this defect was rescued by restoring FRM-3 expression in body muscles (M resc.). mIPSCs were recorded from adult body wall muscles. Representative traces (E), mean amplitude (F) and mean frequency (G) are shown. (H) The function of total surface UNC-49 receptors was unaltered in frm-3 mutants. Muscimol-activated currents were recorded from adult body muscles. Mean peak currents are shown. Values that differ significantly are indicated (***, p < 0.001; **, p < 0.01; ns, not significant). The number of animals analysed is indicated for each genotype. Error bars indicate SEM.
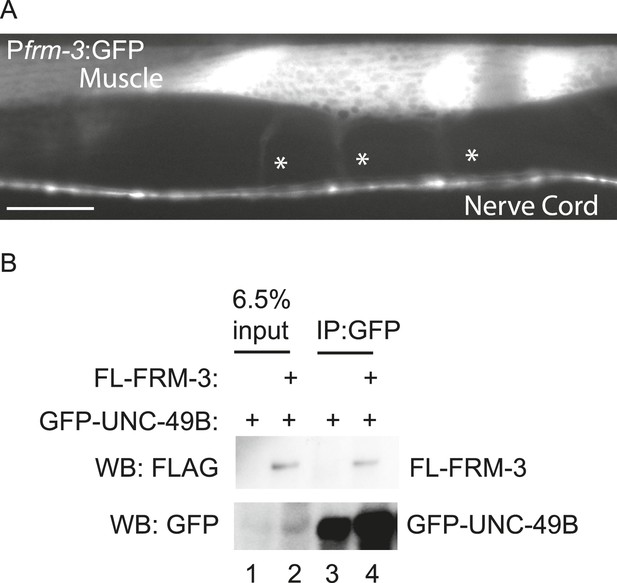
FRM-3 is expressed in body muscles and binds to UNC-49B.
(A) Transgenes containing the frm-3 promoters express GFP in body muscles. Muscle arms are indicated by the asterisks. Scale bar, 25 μm. (B) FLAG-tagged FRM-3 and GFP-UNC-49B co-immunoprecipitate from worm extracts. FLAG-FRM-3 was immunoprecipitated from worm membrane extracts and bound proteins were analyzed by immunoblotting with FLAG (top) and GFP (bottom) antibodies.
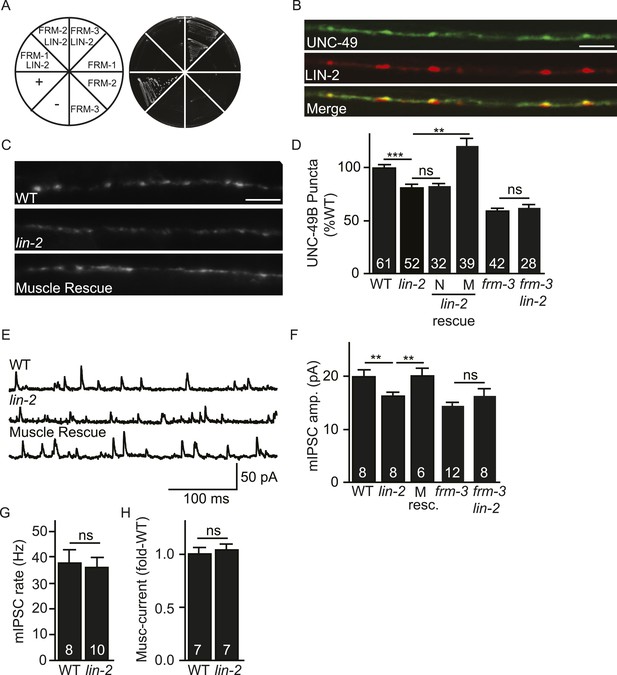
LIN-2A/CASK binds FRM-3 and is required for UNC-49 synaptic targeting.
(A) FRM-3′s ERM domain binds LIN-2A in yeast 2-hybrid assays. Growth of Y2HGold cells on selective media (–Trp/-Leu/-His/-Ade) is shown. Yeast cells were transformed with vectors expressing the indicated fusion proteins. Positive (+, pGBKT7-53 and pGADT7-T) and negative (−, pGBKT7-Lam and pGADT7-T) controls are indicated. ERM domains derived from FRM-1, FRM-2 and FRM-3 were tested for interaction with LIN-2A. (B) Muscle expressed GFP-UNC-49B (Green) and LIN-2::mCherry (Red) are co-localized in the nerve cord. A representative image is shown (scale bar 5 μm). (C, D) GFP-UNC-49B puncta fluorescence in the nerve cord was decreased in lin-2 mutants. This defect was rescued by transgenes expressing LIN-2A in body muscles (M) but not by those expressed in GABergic neurons (N). Representative images (C, scale bar 5 μm) and mean puncta intensity (D) are shown. (E–G) mIPSC amplitude was reduced in lin-2 mutants and this defect was rescued by a transgene expressing LIN-2 in body muscle (M resc). mIPSCs were recorded from adult body muscles. Representative traces (E), mean amplitude (F), and mean frequency (G) are shown. (H) Muscimol-activated currents in adult body muscles were unaffected in lin-2 mutants, indicating that the function of total surface UNC-49 receptors was unaltered. Mean peak currents are shown. lin-2 and frm-3 mutations did not have additive effects on UNC-49B puncta fluorescence (D) or mIPSC amplitudes (F) in double mutants. Values that differ significantly are indicated (***, p < 0.001; **, p < 0.01; ns, not significant). The number of animals analysed is indicated for each genotype. Error bars indicate SEM.
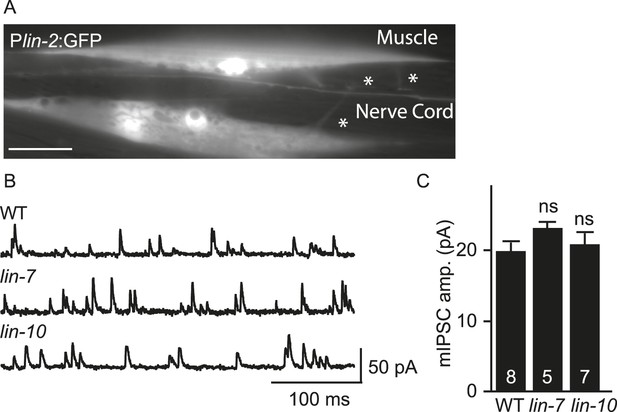
mIPSC amplitudes were unaltered in lin-7 and lin-10 mutants.
(A) Transgenes containing the lin-2 promoters express GFP in body muscles. Muscle arms are indicated by asterisks. Scale bar, 25 μm. (B, C) mIPSCs were recorded from adult body wall muscles of lin-7 and lin-10 mutants. Representative traces (B) and mean mIPSC amplitude (C) are shown. No significant differences were observed. The number of animals analysed is indicated for each genotype. Error bars indicate SEM.
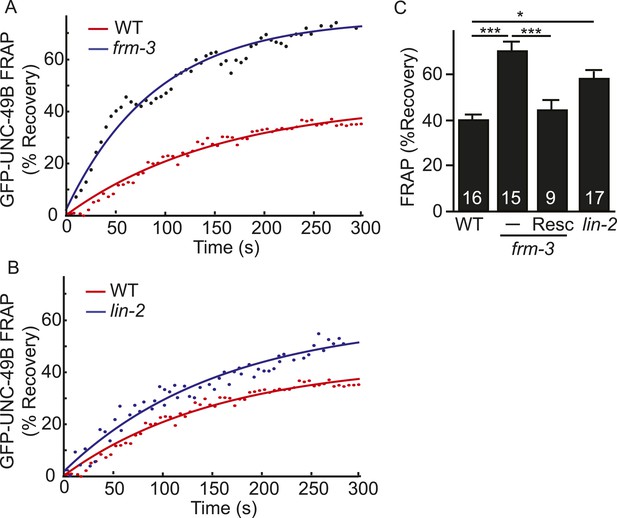
LIN-2A and FRM-3 stabilize immobile UNC-49B receptors at synapses.
Mobility of synaptic GFP-UNC-49B was analyzed by FRAP. Representative scatter plots of fluorescence recovery (solid lines indicate single exponential fits) (A, B) and summary data (C) are shown. Fluorescence recovery was increased in frm-3 and lin-2 mutants, indicating increased mobility of synaptic UNC-49B. The frm-3 mutant FRAP defect was rescued by a transgene expressing FRM-3 in body muscles (Resc). Values that differ significantly are indicated (***, p < 0.001; *, p < 0.05). The number of animals analyzed is indicated for each genotype. Error bars indicate SEM.
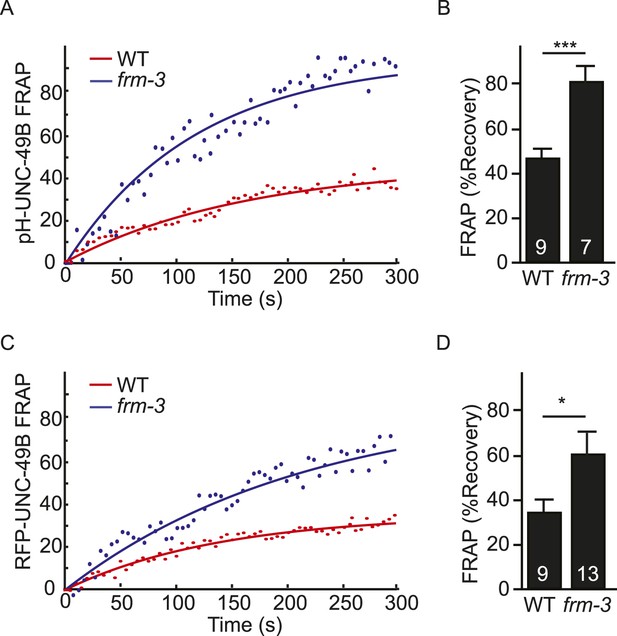
FRAP analysis of pH-UNC-49B and single copy RFP-UNC-49B in frm-3 mutants.
To assess the mobility of surface UNC-49B receptors at synapses, we analyzed FRAP of pHluorin-tagged UNC-49B (pH-UNC-49B) and of an RFP-UNC-49B single copy transgene (krSi2). FRAP of pH-UNC-49B (A, B) and RFP-UNC-49B (C, D) were both significantly increased in frm-3 mutants. Representative scatter plots of fluorescence recovery and single exponential fits (solid lines) (A, C), and summary data (B, D) are shown. The number of animals analysed is indicated for each genotype. Error bars indicate SEM. Values that differ significantly from WT controls are indicated (***, p < 0.001; *, p < 0.05).
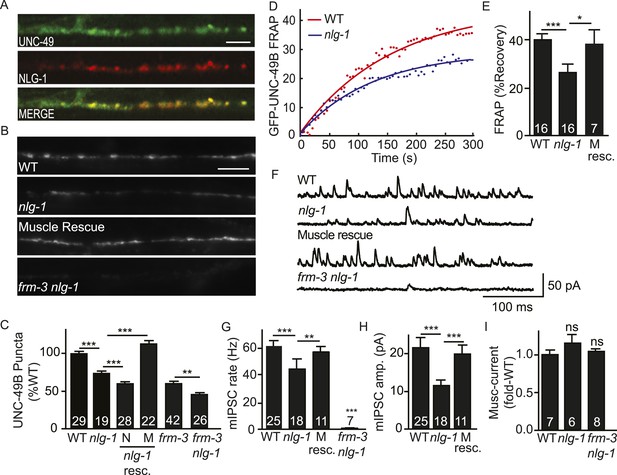
NLG-1 stabilizes mobile UNC-49B at synapses.
(A) Muscle expressed GFP-UNC-49B (Green) and NLG-1::mCherry (Red) are co-localized in the nerve cord. A representative image is shown (scale bar 5 μm). (B, C) GFP-UNC-49B synaptic abundance was decreased in nlg-1 mutants and this defect was rescued by transgenes expressing NLG-1 in body muscles (M) but not by those expressed in motor neurons (N). Representative images (B, scale bar 5 μm) and mean puncta intensity (C) are shown. (D, E) Mobility of synaptic GFP-UNC-49B was analyzed by FRAP. Representative scatter plots of fluorescence recovery and single exponential fits (solid lines) (D) and summary data (E) are shown. Fluorescence recovery was decreased in nlg-1 mutants, indicating that synaptic UNC-49B mobility was decreased. The nlg-1 mutant FRAP defect was rescued by a transgene expressing NLG-1 in body muscles (M Resc) (E). (F–H) mIPSC amplitude was reduced in nlg-1 mutants and this defect was rescued by a transgene expressing NLG-1 in body muscle (M resc.). mIPSCs were recorded from adult body muscles. Representative traces (F), mean frequency (G), and mean amplitude (H) are shown. (I) Muscimol-evoked currents (mean peak amplitude) was unaltered in nlg-1 and in frm-3 nlg-1 double mutants. nlg-1 and frm-3 mutations had additive effects on UNC-49B puncta fluorescence (B, C) and mIPSCs (F) in double mutants. The number of animals analyzed is indicated for each genotype. Error bars indicate SEM. Values that differ significantly are indicated (***, p < 0.001; **, p < 0.01; *, p < 0.05; ns, not significant).
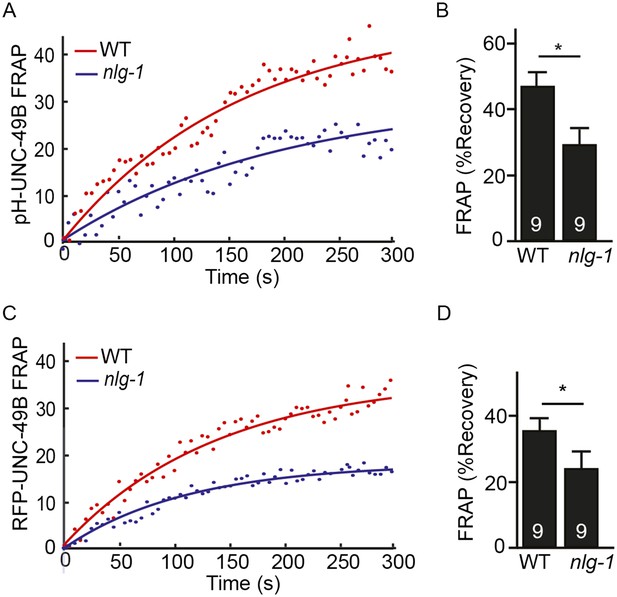
FRAP analysis of pH-UNC-49B and single copy RFP-UNC-49B in nlg-1 mutants.
To assess the mobility of surface UNC-49B receptors at synapses, we analyzed FRAP of pHluorin-tagged UNC-49B (pH-UNC-49B) and of an RFP-UNC-49B single copy transgene (krSi2). FRAP of pH-UNC-49B (A, B) and RFP-UNC-49B (C, D) was significantly decreased in nlg-1 mutants. Representative scatter plots of fluorescence recovery and single exponential fits (solid lines) (A, C), and summary data (B, D) are shown. The number of animals analysed is indicated for each genotype. Error bars indicate SEM. Values that differ significantly from WT controls are indicated (*, p < 0.05).
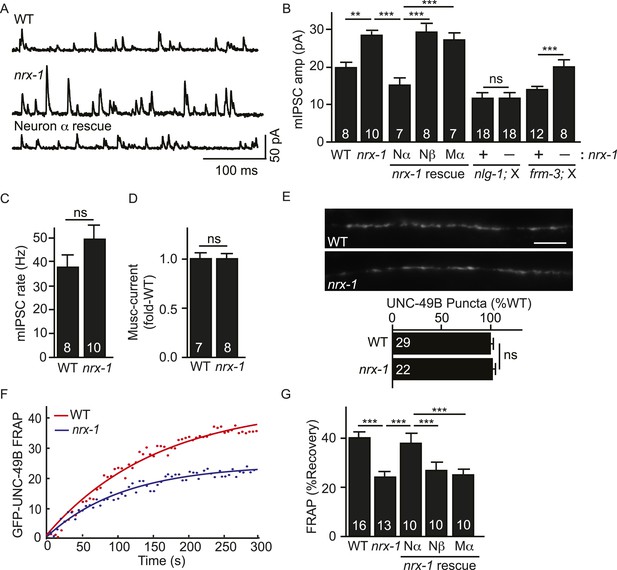
Pre-synaptic NRX-1α inhibits immobilization of synaptic UNC-49B.
(A, B) Mutations inactivating nrx-1 increased mIPSC amplitude and this defect was rescued by transgenes expressing NRX-1α in motor neurons (Nα) but not those expressing NRX-1β (Nβ). Transgenes expressing NRX-1α in body muscles (Mα) lacked rescuing activity. mIPSCs were recorded from adult body muscles. Representative traces (A), mean amplitude (B) and mean frequency (C) are shown. The effect of nrx-1 mutations on mIPSC amplitudes was eliminated in nrx-1;nlg-1 double mutants but was unaffected in nrx-1; frm-3 double mutants (B). (D) Muscimol-evoked currents (mean peak amplitude) was unaffected in nrx-1 mutants. (E) GFP-UNC-49B synaptic abundance was unaltered in nrx-1 mutants Representative images (top, scale bar 5 μm) and mean puncta intensity (below) are shown. (F, G) FRAP analysis suggests that mobility of synaptic GFP-UNC-49B was decreased in nrx-1 mutants. This FRAP defect was rescued by transgenes expressing NRX-1α in motor neurons (Nα) but not those expressing NRX-1β (Nβ). Transgenes expressing NRX-1α in body muscles (Mα) lacked rescuing activity. Representative scatter plots of fluorescence recovery and single exponential fits (solid lines) (F) and summary data (G) are shown. The number of animals analyzed is indicated for each genotype. Error bars indicate SEM. Values that differ significantly are indicated (***, p < 0.001; **, p < 0.01; ns, not significant).
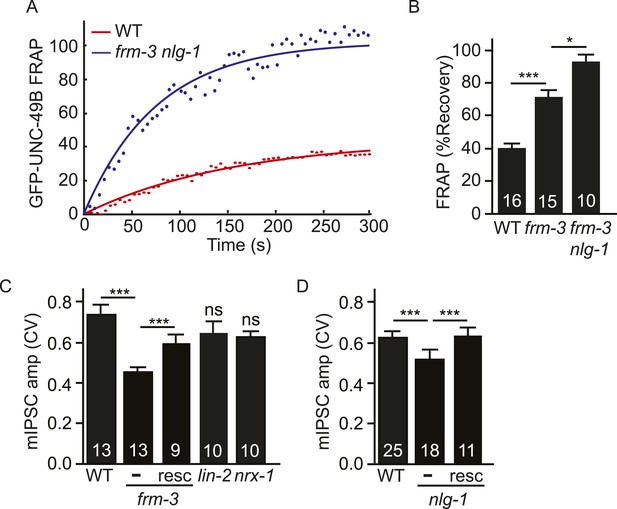
Both UNC-49 receptor pools contribute to post-synaptic responses.
(A, B) FRAP analysis suggests that the immobile pool of synaptic GFP-UNC-49B was eliminated in frm-3 nlg-1 double mutants. Representative scatter plots of fluorescence recovery and single exponential fits (solid lines) (A) and summary data (B) are shown. (C, D) The FRM-3 and NLG-1 scaffolds increase the diversity of quantal responses. CV of mIPSC amplitudes are shown for the indicated genotypes. CV was significantly decreased in frm-3 (C) and in nlg-1 (D) mutants. The frm-3 and nlg-1 CV defects were rescued by transgenes restoring expression of the corresponding genes in body muscles (resc). CV was not significantly altered in lin-2 and nrx-1 mutants (C). The number of animals analyzed is indicated for each genotype. Error bars indicate SEM. Values that differ significantly are indicated (***, p < 0.001; *, p < 0.05; ns, not significant).





