Auxiliary subunits of the CKAMP family differentially modulate AMPA receptor properties
Figures
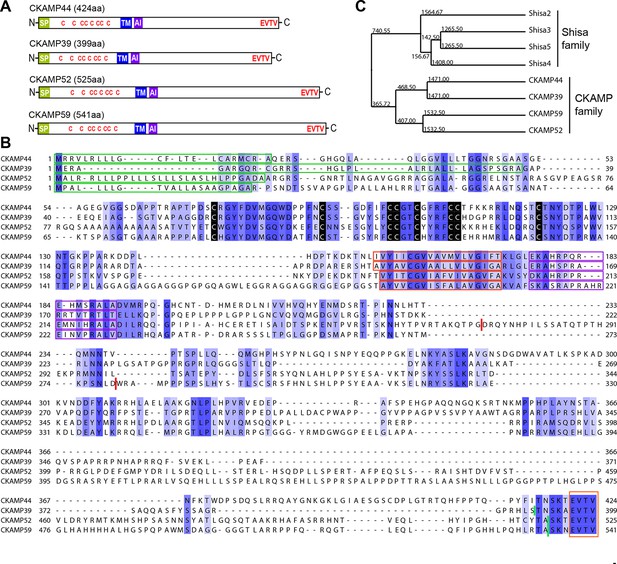
Identification and molecular characterization of CKAMP proteins.
(A) Schematic drawing of CKAMP proteins, depicting the signal peptide (SP), cysteines (C) of the cystine knot, transmembrane domain (TM) and PDZ type II motif (EVTV). ‘AI’ indicates “AMPAR interacting” region, based on the GluA1 binding co-IP experiments from Khodosevich et al. (2014). (B) Protein sequence alignment of mouse CKAMP39, CKAMP44, CKAMP52 and CKAMP59. Amino acids marked in blue are similar or identical among CKAMP family members. Intensity of the blue color indicates the degree of similarity, with identical amino acid positions in the protein sequence highlighted by the most intense color. Green rectangles outline the predicted signal peptides, red rectangles—the predicted transmembrane domains, purple rectangles - putative AMPAR interacting regions and orange rectangle—the PDZ type II motif. Positions of cysteines belonging to the cystine-knot motif are indicated in black. Thick red lines in position 274 for CKAMP52 and 280 for CKAMP59 indicate insertion sites for additional aa sequences that are encoded by alternatively spliced exon 3 and exon 4, respectively. Thick green lines nearby C-termini indicate positions for flag-tag insertions. (C) Phylogenetic analysis of Shisa and CKAMP proteins, based on their protein sequence (average distance tree).
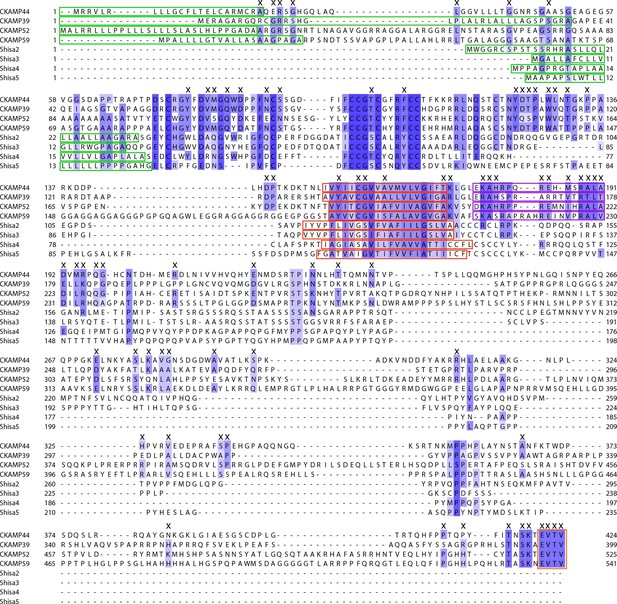
Comparison of Shisa proteins and CKAMPs.
Alignment of protein sequences that correspond to Shisa and CKAMP genes. Note that Shisa1 was originally described for Xenopus laevis oocytes and does not have a mouse homolog. Intensity of the blue color indicates the degree of similarity, with identical amino acid positions in the protein sequence highlighted by the most intense color. “X” labels those amino acids that are conserved in the CKAMP, but not in Shisa family. Green rectangles outline the predicted signal peptides, red rectangles—the predicted transmembrane domains, purple rectangles—putative AMPAR interacting regions and orange rectangle—the PDZ type II motif.
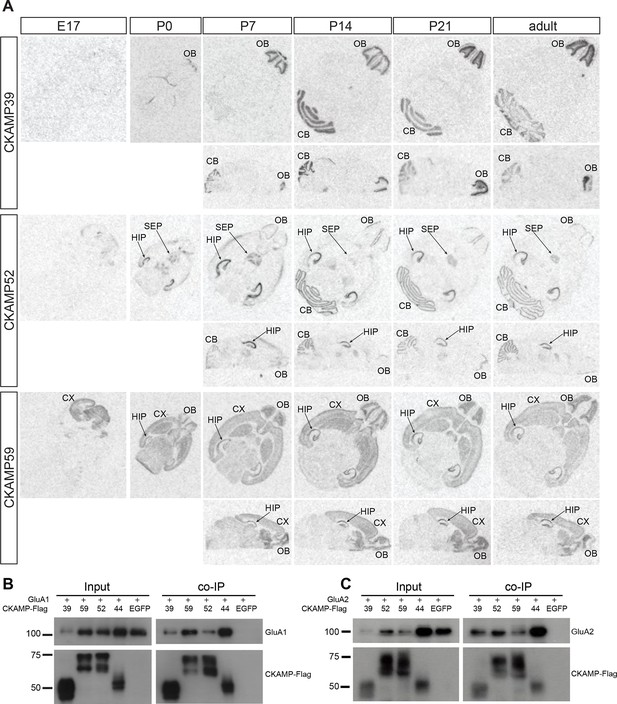
Expression pattern of CKAMPs and their interaction with AMPARs.
(A) Transcription of CKAMP genes as visualized by in situ hybridization of brain sections obtained from mice at different developmental ages. Abbreviations: CB - cerebellum, CX - cortex, HIP - hippocampus, OB - olfactory bulb, SEP - septum. (B) and (C) All members of the CKAMP family interact with GluA1 and GluA2, respectively. HEK293/T17 cells were transfected with GluA1 (B) or GluA2 (C) together with one of the indicated flag-tagged CKAMPs or with EGFP as a control. Proteins were immunoprecipitated using anti-flag antibody. All flag-tagged CKAMPs, but not EGFP, co-precipitated GluA1 (B) or GluA2 (C) from the total protein fraction. Input corresponds to ~9% of GluA1 or ~3% of GluA2 co-immunoprecipitation experiment.

Analysis of splice isoforms for CKAMP family members.
Splice variants for CKAMP52 and CKAMP59 (note that CKAMP39 has only one splice variant). Exons are numbered below the scheme of the spliced mRNA. For both CKAMPs, at least two splice isoforms are present in the brain, with or without the 3rd (96 nt) and 4th (51 nt) exon for CKAMP52 and CKAMP59, respectively (marked in red).
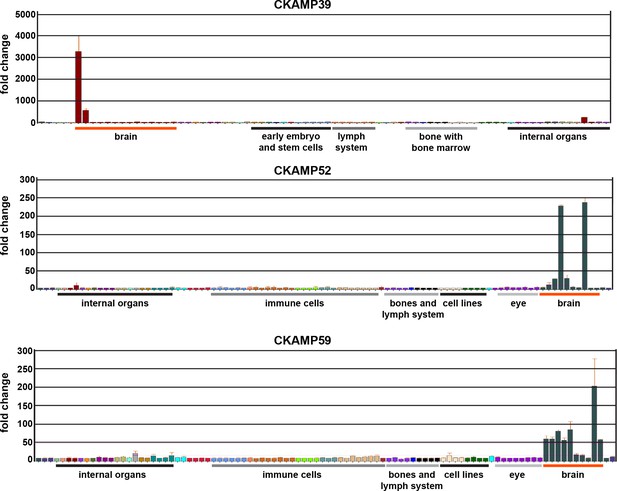
Analysis of expression for CKAMP family members.
Expression of novel CKAMP family members in different mouse tissues according to the BioGPS database. Note that a significant expression of CKAMP39 was found only in the olfactory bulb and cerebellum, which corresponds to our in situ hybridization data in Figure 2A.
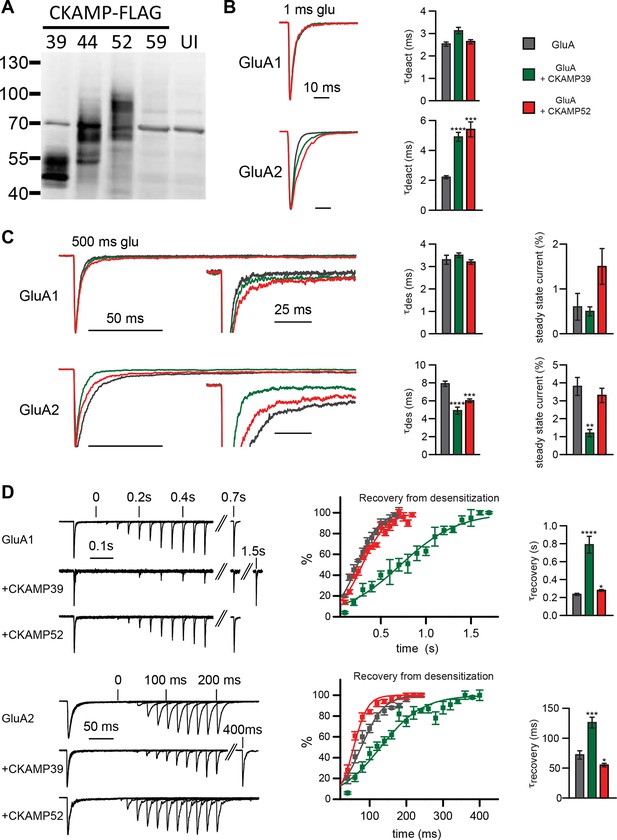
Modulation of AMPAR-mediated currents in Xenopus laevis oocytes.
(A) Western blot analysis on flag-tagged CKAMPs in Xenopus laevis oocytes. CKAMP39, CKAMP44, CKAMP52 & CKAMP59 where injected at 1, 3, 5 & 10 ng/oocyte, respectively (UI-un-injected oocytes). (B) Deactivation rate (τdeact), (C) desensitization rate (τdes) and steady-state amplitude and (D) weighted time constant of recovery from desensitization (τrecovery) of GluA1- and GluA2(Q)-mediated currents. Deactivation and desensitization were tested by application of 10 mM glutamate for 1 ms and 500 ms, respectively, onto macropatches of oocytes. τrecovery was estimated with application of two 100 ms glutamate pulses with different inter-pulse intervals. Example currents are shown on the left of the quantification (mean ± SEM).
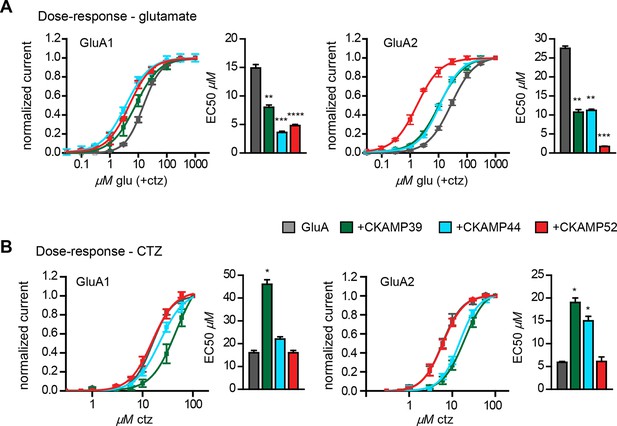
Influence of CKAMP proteins on glutamate and CTZ potency.
(A) Glutamate potency for GluA1 or GluA2(Q) expressed along with CKAMP39, CKAMP44 or CKAMP52 in Xenopus laevis oocytes. Responses to increasing concentration of glutamate were recorded in the presence of 0.1 mM CTZ applied 10 s before and during glutamate application to reach equilibrium steady-state currents. Responses were normalized to the response obtained for 1mM glutamate. (B) CTZ potency for GluA1 or GluA2(Q) expressed along with CKAMP39, CKAMP44 or CKAMP52 in Xenopus laevis oocytes. Responses to 1 mM glutamate were recorded in the presence of increasing concentrations of CTZ after a 10 s incubation of the oocyte with the respective CTZ concentration without glutamate. Current responses were normalized to the response obtained with 0.1 mM CTZ after subtraction of the current induced by 1 mM glutamate without CTZ. The respective EC50 values (mean ± SEM) were calculated from fits to data obtained from individual oocytes.
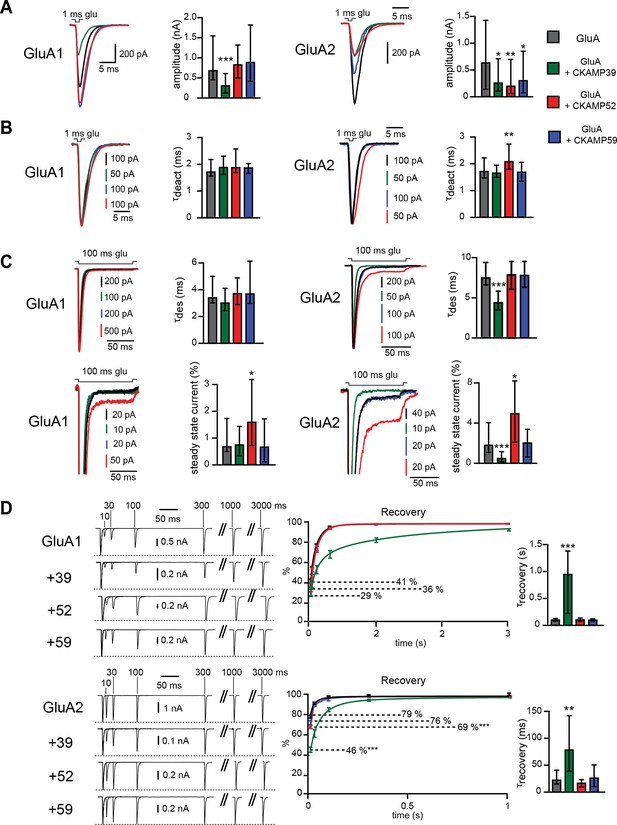
Modulation of AMPAR-mediated currents in HEK293/T17 cells.
(A) Peak current amplitude, (B) deactivation rate (τdeact), (C) desensitization rate (τdes) and steady-state amplitude and (D) τrecovery of GluA1- and GluA2(Q)-mediated currents. Deactivation and desensitization were tested by application of 1 mM glutamate for 1 ms and 100 ms, respectively. τrecovery was analyzed with application of two 1 ms glutamate pulses with different inter-pulse intervals. Example currents are shown on the left of the quantification (median ± IQR).
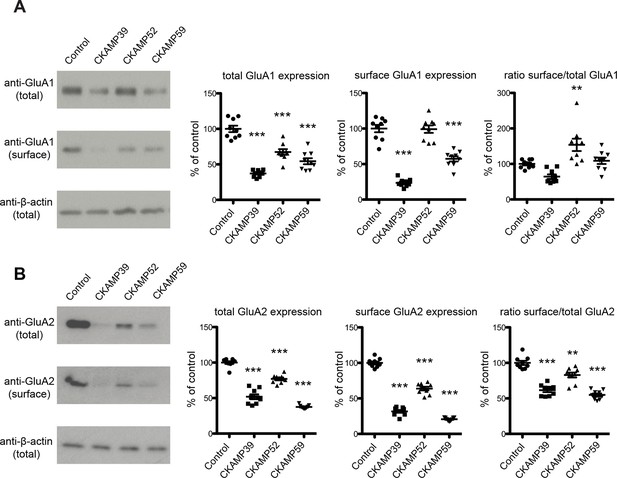
Analysis of total and surface AMPAR expression in HEK293/T17 cells.
Example Western blot images (left) and quantification (right) of total AMPAR, surface AMPAR, and β-actin. GluA1 (A) or GluA2 (B) was expressed in HEK293/T17 cells along with CKAMP family members or a membrane-bound GFP as a control. Cell surface protein was biotinylated and separated from total protein using affinity gel. Results are presented as a percent of control values (Mean ± SEM).
Additional files
-
Supplementary file 1
- https://doi.org/10.7554/eLife.09693.012





