Fat body-derived cytokine Upd2 controls disciplined migration of tracheal stem cells in Drosophila
Figures
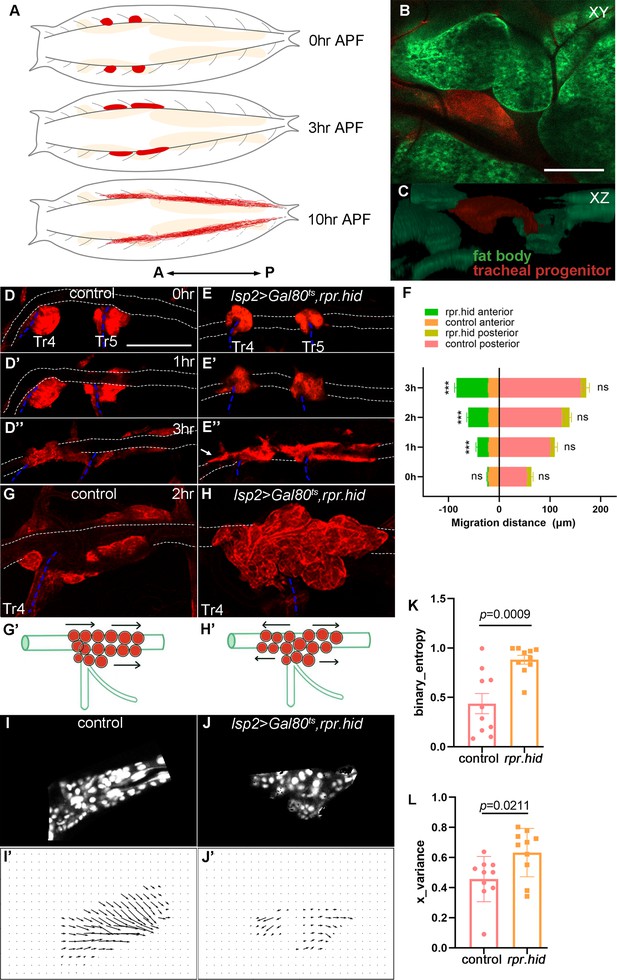
Fat body affects disciplined migration of tracheal progenitors.
(A) Schematic cartoon showing the migration of tracheal progenitors (red) and degenerative tracheal branches (dashed gray lines) in pupae. Fat body is shown in beige. Arrows denote anterior–posterior (A–P) axis. Frontal section (B) and sagittal view (C) showing the relative position of fat body and tracheal progenitors. (D–J’) Migration of tracheal progenitors in control and fat body perturbation flies. (D–D’’) Migration of tracheal progenitors (red) upward from transverse connective (blue dashed lines) and along the dorsal trunk (white dotted lines) at 0 hr APF (D), 1 hr APF (D’), and 3 hr APF (D’’). Bidirectional movement of tracheal progenitors in fat body-depleted (lsp2>rpr.hid) flies. 0 hr APF (E), 1 hr APF (E’), and 3 hr APF (E’’). Arrows point to anterior movement of tracheal progenitors. (F) Bar graph showing the migration distance of tracheal progenitors. The top chart of column represents the migration distance of anterior-most stem cells, and the lower chart of column represents the migration distance of posterior-most stem cells. Error bars represent SEM, n = 6. (G, G’) The distribution of progenitors at 2 hr APF. (H, H’’) The distribution of progenitors in fat body-depleted flies at 2 hr APF. (I–J’) Computer simulation depicting trajectories of progenitor migration. (I, J) Confocal images of tracheal progenitors. (I’, J’) Vectors of progenitor migration. (K) Bar graph plots the binary entropy that represents the disorderedness of migration direction of tracheal progenitors. Error bars represent SEM, n = 10. (L) The Bernoulli random variable X showing optic flow distribution of the binarized directions in each group. Error bars represent SEM, n = 10. N.S. indicates not significant. Scale bar: 100 μm (B, C, G), 200 μm (D–E’’). Genotypes: (B, C) UAS-mCD8-GFP/+; lsp2-Gal4,P[B123]-RFP-moe/+; (D–D’’, G, G’) Gal80ts/+;lsp2-Gal4,P[B123]-RFP-moe/+; (E–E’’, H, H’) UAS-rpr-hid/+;Gal80ts/+;lsp2-Gal4,P[B123]-RFP-moe/+.
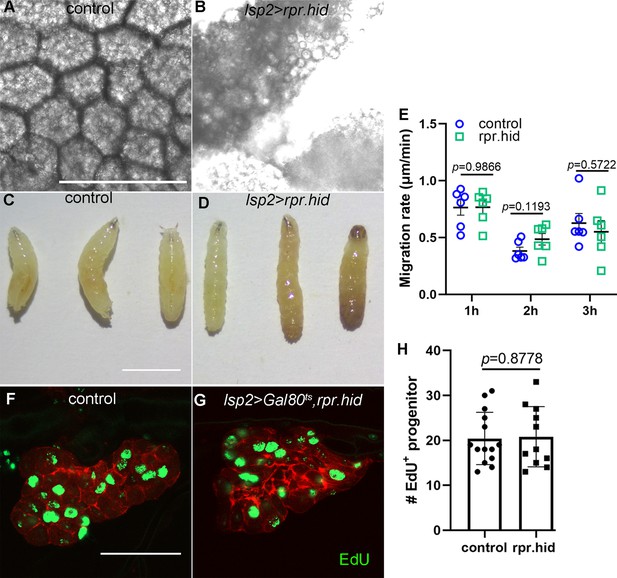
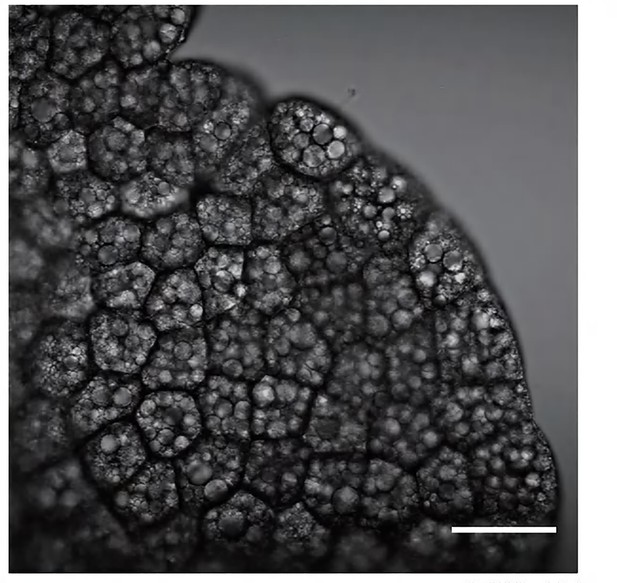
Genetic perturbation of fat body by expression of pro-apoptotic genes, rpr and hid.
Confocal images of larval fat body in control (A) and lsp2>rpr.hid (B) animals. Microscopic pictures of control (C) and lsp2>rpr.hid larvae (D). (E) Scatter plot showing migration velocity of tracheal progenitors in control and lsp2>rpr.hid animals. Error bars represent SEM, n = 6. The incorporation of EdU in the tracheal progenitors of control (F) and lsp2>rpr.hid animals (G). (H) Bar graph depicting the number of EdU incorporation. Error bars represent SEM, n = 11. Scale bar: 100 μm (A, B), 20 μm (C, D), 50 μm (F, G). Genotypes: (A, C, F) lsp2-Gal4,P[B123]-RFP-moe/+; (B, D) UAS-rpr.hid/+;lsp2-Gal4,P[B123]-RFP-moe/+; (G) UAS-rpr.hid/+;Gal80ts/+;lsp2-Gal4,P[B123]-RFP-moe/+.
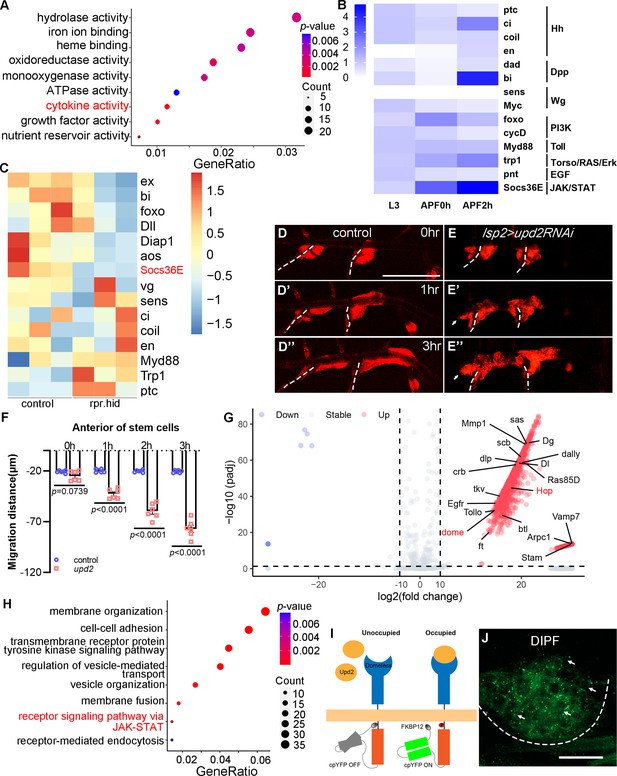
Dependence of tracheal progenitors on cytokines from fat body.
(A) Top functional clusters among the differentially expressed genes of progenitors between control and fat body-depleted pupae. Gene ratio refers to the proportion of genes in a dataset that are associated with a particular biological process, function, or pathway. Count indicates the number of genes from an input gene list that are associated with a specific GO term. (B) Heatmap depicting expression levels of principal target genes of signaling pathways in L3 larvae, 0 hr APF pupae and 2 hr APF pupae. (C) Heatmap showing the differential expression of target genes of signaling pathways between control and fat body-depleted pupae. Migration of tracheal progenitors along the dorsal trunk at 0 hr APF (D), 1 hr APF (D’), and 3 hr APF (D’’). The white dashed line shows transverse connective. (E–E’’). Migration of tracheal progenitors in upd2RNAi flies. (F) Bar graph plots the migration distance of tracheal progenitors. Error bars represent SEM, n = 6. (G) Volcano plot showing surface proteomics of tracheal epithelium (upregulated genes with tenfold or higher changes in red; downregulated genes with tenfold or higher changes in blue). (H) Top functional classes among the surface proteomics of trachea. (I) Schematic diagram depicting the working principle of the DIPF reporter. (J) The signal of DIPF reporter in tracheal progenitors. The progenitors are outlined by dashed lines. N.S. indicates not significant. Scale bar: 200 μm (D–E’’), 50 μm (J). Genotypes: (A, C) lsp2-Gal4,P[B123]-RFP-moe/+ for control, UAS-rpr-hid/+;Gal80ts/+;lsp2-Gal4,P[B123]-RFP-moe/+; (B) P[B123]-RFP-moe/+. (D–D’’) lsp2-Gal4,P[B123]-RFP-moe/+; (E–E’’) lsp2-Gal4,P[B123]-RFP-moe/UAS-upd2RNAi; (J) btl-Gal4/UAS-DIPF.
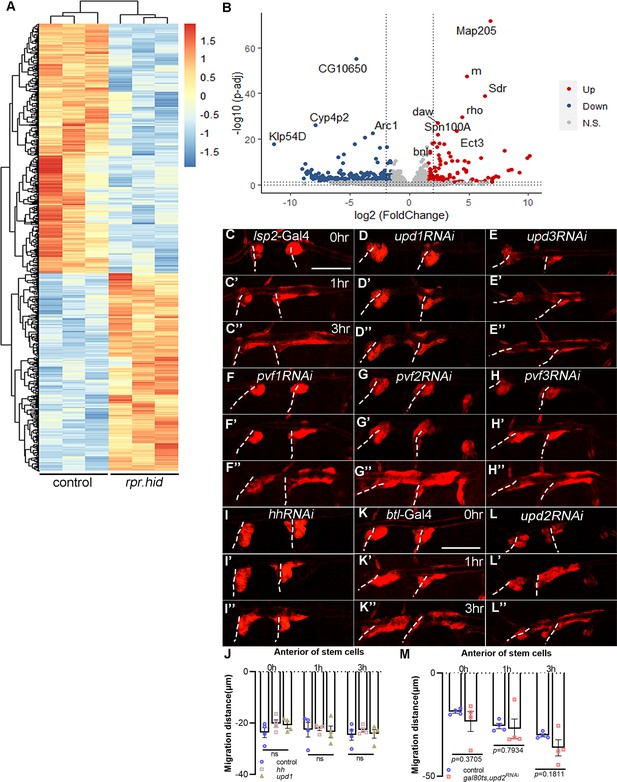
Identification of genes that contribute to progenitors migration.
(A) Differentially expressed genes in tracheal progenitors of lsp2>rpr.hid pupa compared with control. (B) Volcano plot of RNA-seq showing differentially regulated genes with twofold or higher changes (upregulated genes in red; downregulated genes in blue) in lsp2>rpr.hid compared with control. (C–I’’) The migration of progenitors at 0 hr APF, 1 hr APF, or 3 hr APF in control (C–C’’) and upd1RNAi (D–D’’), upd3RNAi (E–E’’), pvf1RNAi (F–F’’), pvf2RNAi (G–G’’), pvf3RNAi (H–H’’), and hhRNAi (I–I’’). (J) Bar graph represents the migration distance of anterior movement. Error bars represent SEM, n = 4. (K) The migration of progenitors in control (K–K’’) and btl>upd2 RNAi (L–L’’). (M) Bar graph plots the migration distance of anterior movement. Error bars represent SEM, n = 4. N.S. indicates not significant. Scale bar: 200 μm (C–I’’, K–L’’). Genotypes: (C–C’’) lsp2-Gal4,P[B123]-RFP-moe/+; (D–D’’) lsp2-Gal4,P[B123]-RFP-moe/UAS-upd1RNAi; (E–E’’) lsp2-Gal4,P[B123]-RFP-moe/UAS-upd3RNAi; (F–F’’) lsp2-Gal4,P[B123]-RFP-moe/UAS-pvf1RNAi; (G–G’’) lsp2-Gal4,P[B123]-RFP-moe/UAS-pvf2RNAi; (H–H’’) lsp2-Gal4,P[B123]-RFP-moe/UAS-pvf3RNAi; (I–I’’) lsp2-Gal4,P[B123]-RFP-moe/UAS-hhRNAi; (K–K’’) btl-Gal4/+;P[B123]-RFP-moe/+; (L–L’’) btl-Gal4/+;P[B123]-RFP-moe/UAS-upd2RNAi.

Surface proteomics of Drosophila trachea.
Representative confocal images showing the trachea of btl >CD2-HRP flies after oxidation reaction in the presence of both BXXP and H2O2 (A), BXXP only (B), or H2O2 only (C). (D) The western blot showing the biotinylated proteins labeled by an HRP-catalyzed reaction with both BXXP and H2O2. (E) The confocal image showing the trachea of dome>GFP flies. The expression domain of dome is visualized by dome-Gal4-controlled GFP expression. The progenitors labeled by a progenitor-specific enhancer, P[B123]-RFP-moe are outlined by dashed lines. The overlapping expression of GFP and P[B123]-RFP-moe. The expression of DIPF in fat body (F) and salivary gland (G). Scale bar: 50 μm (E–H). Genotypes: (A–C) btl-Gal4/UAS-CD2-HRP; (E) dome-Gal4/+;UAS-GFP/P[B123]-RFP-moe; (F, G) tub-Gal4/UAS-DIPF.
-
Figure 2—figure supplement 2—source data 1
Original files for western blot analysis displayed in Figure 2—figure supplement 2D.
- https://cdn.elifesciences.org/articles/100037/elife-100037-fig2-figsupp2-data1-v1.zip
-
Figure 2—figure supplement 2—source data 2
PDF file containing original western blots for Figure 2—figure supplement 2D, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100037/elife-100037-fig2-figsupp2-data2-v1.zip
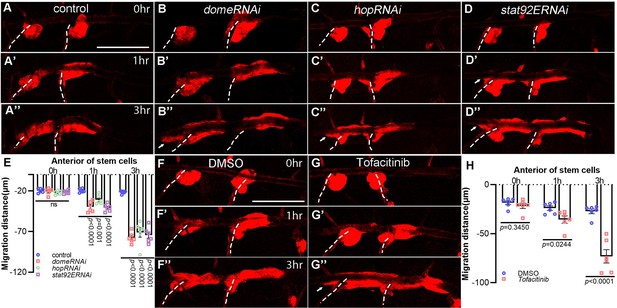
JAK/STAT pathway is required for the discipline of tracheal progenitor migration.
(A–D’’) Migration of tracheal progenitors along the dorsal trunk at 0 hr APF, 1 hr APF, and 3 hr APF. The white dashed line shows transverse connective. The progenitors of control (A–A’’), domeRNAi (B–B’’), hopRNAi (C–C’’), and stat92ERNAi (D–D’’) flies. (E) Bar graph showing migration distance of progenitors. Error bars represent SEM, n = 6. (F–G’’) JAK inhibition causes bidirectional movement of progenitors. Migration of tracheal progenitors in the absence (DMSO-fed) (F–F’’) or in the presence of Tofacinib (JAK inhibitor) (G–G’’). (H) Bar graph showing the distance of anterior movement. Error bars represent SEM, n = 6. Scale bar: 200 μm (A–D’’, F–G’’). Genotypes: (A–A’’, F–G’’) btl-Gal4/+;P[B123]-RFP-moe/+; (B–B’’) btl-Gal4/+;P[B123]-RFP-moe/UAS-domeRNAi; (C–C’’) btl-Gal4/+;P[B123]-RFP-moe/UAS-hopRNAi; (D–D’’) btl-Gal4/+;P[B123]-RFP-moe/UAS-stat92ERNAi.
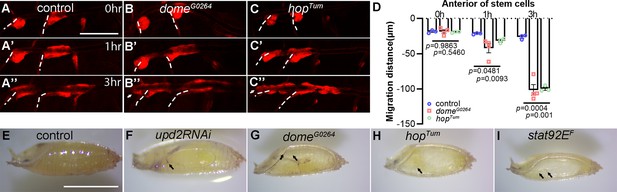
The roles of JAK/STAT pathway in tracheal development.
(A–E) Migration of tracheal progenitors along the dorsal trunk at 0 hr APF, 1 hr APF, and 3 hr APF. The white dashed line shows transverse connective. The progenitors of control (A–A’’), dome mutants (B–B’’), hop mutants (C–C’’). (D) Bar graphs represent migration distance of anterior movement. Error bars represent SEM, n ≥ 3. N.S. indicates not significant. The pupal trachea in control (E), upd2 RNAi (F), domeG0264 (G), hoptum (H), and stat92EF (I). Scale bar: 200 μm (A–E), 20 mm (K–L’’). Genotypes: (A, E) lsp2-Gal4,P[B123]-RFP-moe/+ (control); (B) domeG0264; lsp2-Gal4,P[B123]-RFP-moe/+; (C) hoptum; lsp2-Gal4,P[B123]-RFP-moe/+; (F) lsp2-Gal4,P[B123]-RFP-moe/upd2RNAi. (G) domeG0264. (H) hoptum. (I) stat92EF.
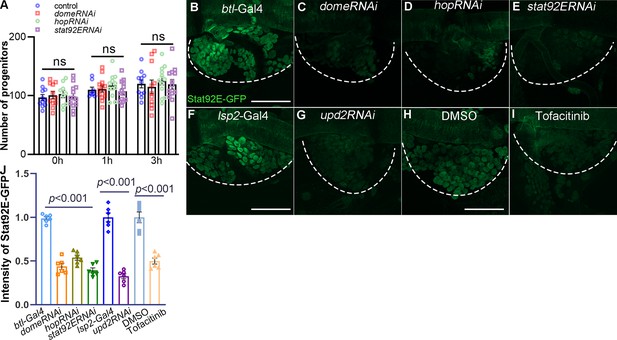
The dependence of tracheal progenitors on JAK/STAT pathway.
(A) Statistics of the number of progenitor cells. Error bars represent SEM, n ≥ 9. N.S. indicates not significant. The expression of Stat92E-GFP in tracheal progenitors of control (B), domeRNAi (C), hopRNAi (D), and stat92ERNAi (E) flies. The progenitors are outlined by dashed lines. The expression of Stat92E-GFP in tracheal progenitors of control (F) or upd2RNAi flies (G). The expression of Stat92E-GFP in tracheal progenitors of DMSO-fed control, (H) or Tofacitinib-treated (I) flies. Dashed lines outline tracheal progenitors. (J) Bar graph plots relative intensity of Stat92E-GFP reporter. Error bars represent SEM, n = 6. N.S. indicates not significant. Scale bar: 50 μm (B–I). Genotypes: (A) btl-Gal4/+;P[B123]-RFP-moe/+(control); btl-Gal4/+;P[B123]-RFP-moe/UAS-domeRNAi; btl-Gal4/+;P[B123]-RFP-moe/UAS-hopRNAi; btl-Gal4/+;P[B123]-RFP-moe/UAS-stat92ERNAi; (B) btl-Gal4,Stat92E-GFP/+; (C) btl-Gal4,Stat92E-GFP/+;UAS-domeRNAi/+; (D) btl-Gal4,Stat92E-GFP/+;UAS-hopRNAi/+; (E) btl-Gal4,Stat92E-GFP/+;UAS-Stat92ERNAi/+; (F, H, I) Stat92E-GFP/+; lsp2-Gal4/+; (G) Stat92E-GFP/+;lsp2-Gal4/UAS-upd2RNAi.
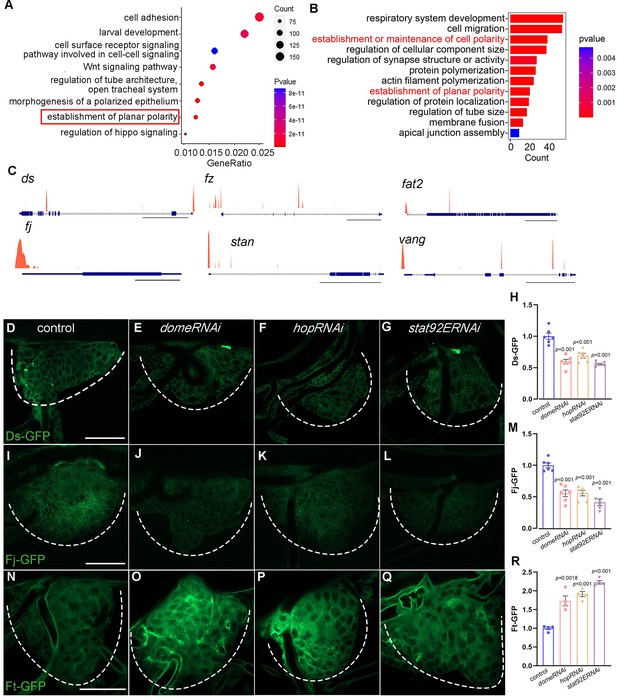
Identification of gene targets of Stat92E in Drosophila tracheal progenitors.
(A) Bubble plot represents the top functional clusters among gene targets. The establishment of planar polarity denoted in red solid box is identified with high enrichment score. (B) Top functional classes among the differentially expressed genes in larval–pupal transition. (C) ChIP-seq peaks at loci regulated by Stat92E. Scale bar: 20 kb (ds, fz, stan), 5 kb (fat2, vg), 1 kb (fj). (D–H) Validation of gene targets of Stat92E ChIP-seq. The expression of Ds-GFP in the tracheal progenitors of control (D), domeRNAi (E), hopRNAi (F), and stat92ERNAi (G). The progenitors are outlined by dashed lines. (H) The bar graphs plot the relative level of Ds. Error bars represent SEM, n = 6. (I–M) The expression of Fj in tracheal progenitors. The expression of Fj-GFP in the tracheal progenitors of control (I), domeRNAi (J), hopRNAi (K), and stat92ERNAi (L). Dashed lines outline tracheal progenitors. (M) The bar graphs plot the relative level of Fj. Error bars represent SEM, n = 6. (N–R) The level of Ft-GFP in the tracheal progenitors of control (N), domeRNAi (O), hopRNAi (P), and stat92ERNAi (Q). (R) The bar graphs plot the relative level of Ft. Error bars represent SEM, n = 4. Scale bar: 50 μm (D–G, I–L, N–Q). Genotypes: (D) btl-Gal4,Ds-GFP/+; (E) btl-Gal4,Ds-GFP/+;UAS-domeRNAi/+; (F) btl-Gal4,Ds-GFP/+;UAS-hopRNAi/+; (G) btl-Gal4,Ds-GFP/+;UAS-stat92ERNAi/+; (I) btl-Gal4,Fj-GFP/+; (J) btl-Gal4,Fj-GFP/+;UAS-domeRNAi/+; (K) btl-Gal4,Fj-GFP/+;UAS-hopRNAi/+; (L) btl-Gal4,Fj-GFP/+;UAS-stat92ERNAi/+; (N) btl-Gal4/+;Ft-GFP/+; (O) btl-Gal4/+;Ft-GFP/UAS-domeRNAi; (P) btl-Gal4/+;Ft-GFP/UAS-hopRNAi; (Q) btl-Gal4/+;Ft-GFP/UAS-stat92ERNAi.
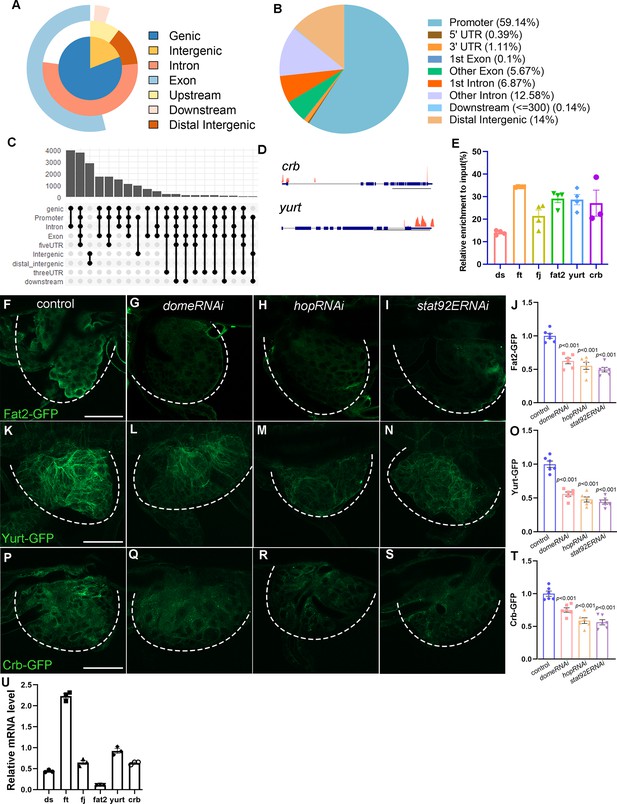
Annotation and analysis of genomic occupancy of Stat92E.
Pie chart (A, B) and histogram (C) depicting the distribution of ChIP-seq peaks relative to the nearby annotated genes. (D) Peaks of Stat92E association in the promotor regions of yurt and crb. Scale bar: 2 kb for yurt, 5 kb for crb. (E) Relative enrichment of JAK/STAT target genes analyzed by ChIP-qPCR. The expression of Fat2-GFP in the tracheal progenitors of control (F), domeRNAi (G), hopRNAi (H), and stat92ERNAi (I). The progenitors are outlined by dashed lines. (J) The bar graphs plot the relative level of Fat2. Error bars represent SEM, n = 6. The expression of Yurt in tracheal progenitors. The expression of Yurt-GFP in the tracheal progenitors of control (K), domeRNAi (L), hopRNAi (M), and stat92ERNAi (N). The progenitors are outlined by dashed lines. (O) The bar graphs plot the relative level of Yurt. Error bars represent SEM, n = 6. The expression of Crb-GFP in the tracheal progenitors of control (P), domeRNAi (Q), hopRNAi (R), and stat92ERNAi (S). The progenitors are outlined by dashed lines. (T) The bar graphs plot the relative level of Crb. Error bars represent SEM, n = 6. (U) The expression of JAK/STAT targets in the trachea of stat92ERNAi flies. Quantitative RT-PCR was performed in triplets to analyze the mRNA level. The expression levels in stat92ERNAi larvae were presented relative to Gal4 control. Scale bar: 50 μm (F–I, K–N, P–S). Genotypes: (F) btl-Gal4,Fat2-GFP/+; (G) btl-Gal4,Fat2-GFP/+;UAS-domeRNAi/+; (H) btl-Gal4,Fat2-GFP/+;UAS-hopRNAi/+; (I) btl-Gal4,Fat2-GFP/+;UAS-stat92ERNAi/+; (J) btl-Gal4/+;Yurt-GFP/+; (K) btl-Gal4/+;Yurt-GFP/UAS-domeRNAi; (L) btl-Gal4/+;Yurt-GFP/UAS-hopRNAi; (M) btl-Gal4/+;Yurt-GFP/UAS-stat92ERNAi; (P) btl-Gal4/+;Crb-GFP/+; (Q) btl-Gal4/+;Crb-GFP/UAS-domeRNAi; (R) btl-Gal4/+;Crb-GFP/UAS-hopRNAi; (S) btl-Gal4/+;Crb-GFP/UAS-stat92ERNAi.

Ds alters the level of Ft.
The level of Ft-GFP in control (A), UAS-ds (B), and dsRNAi (C) flies. (D) Bar graph represents the signal of Ft-GFP. Error bars represent SEM, n = 4. Scale bar: 50 μm (A–C). Genotypes: (A) btl-Gal4/+;Ft-GFP/+; (B) btl-Gal4/+;Ft-GFP/UAS-ds; (C) btl-Gal4/+;Ft-GFP/UAS-dsRNAi.
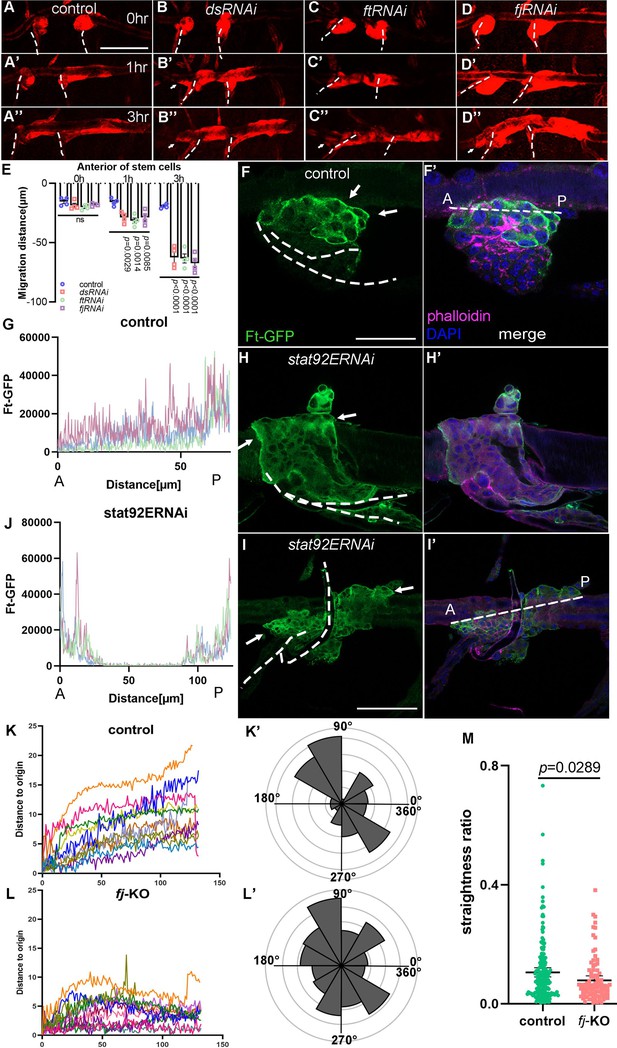
Disciplined migration requires planar cell polarity system.
(A–E) Migration of tracheal progenitors. The migration of progenitors in control (A–A’’), dsRNAi (B–B’’), ftRNAi (C–C’’), and fjRNAi (D–D’’) flies. (E) Bar graph plots the migration distance of anterior movement. Error bars represent SEM, n = 4. Level of Ft in tracheal progenitors of control (F, G) and stat92ERNAi (H–J) flies. The images show progenitors at 1 hr APF (H, H’) and 2 hr APF (I, I’). Ft-GFP (green) (F, H, I), phalloidin (magenta), Hoechst (blue), and merged images (F’, H’, I’). Profile plots showing the level of Ft-GFP in control (G) and stat92ERNAi (J) flies, n = 5. ANOVA test: p < 0.0001. The levels of Ft were measured along the dotted lines in F’ or I’. Anterior (A) and posterior (P). (K) Representative traces plot the migration distance relative to the origin, n = 12. The x-axis represents the number of captured images. Individual frame is captured every 5 min. (K’) Rose plot depicting the direction of cell movement. (L) Representative traces showing the movement of individual fj-KO cells relative to their origin, n = 11. The x-axis represents the number of captured images. Individual frame is captured every 5 min. (L’) Rose plot depicting the movement direction of fj-KO cells. (M) Scatter plots represent the ratio (d/D) of straight-line length displacement (d) relative to the length of the migration track (D) of individual cell. Error bars represent SEM. N.S. indicates not significant. Scale bar: 200 μm (A–D’’), 50 μm (F, F’), 100 μm (H–I’). Genotypes: (A–A’’) btl-Gal4/+;P[B123]-RFP-moe/+; (B–B’’) btl-Gal4/UAS-dsRNAi;P[B123]-RFP-moe/+; (C–C’’) btl-Gal4/+;P[B123]-RFP-moe/UAS-ftRNAi; (D–D’’) btl-Gal4/UAS-fjRNAi;P[B123]-RFP-moe/+; (F, F’) btl-Gal4/+;Ft-GFP/+; (H, H’, I, I’) btl-Gal4/+;Ft-GFP/UAS-stat92ERNAi.
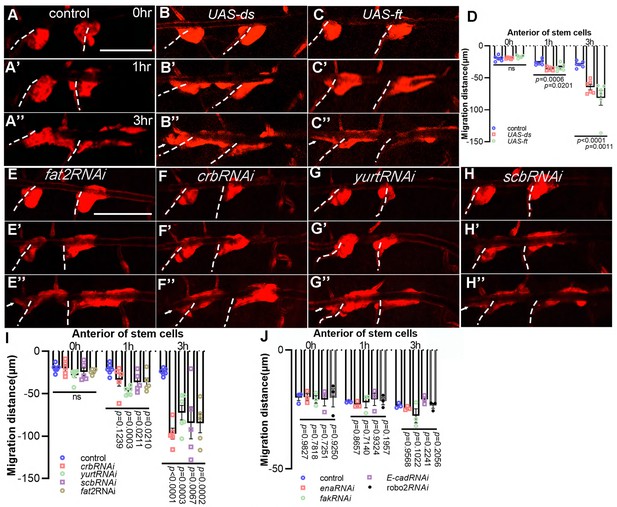
Apico-basal polarity proteins contribute to the migration polarity of progenitors.
The migration of progenitors at 0 hr APF (A), 1 hr APF (A’), 3 hr APF (A’’) in control (A–A’’), UAS-ds (B–B’’), UAS-ft (C–C’’), fat2RNAi (E–E’’), crbRNAi (F–F’’), yurtRNAi (G–G’’) and scbRNAi (H–H’’). (D, I, J) Bar graph plots the migration distance of anterior movement. Error bars represent SEM, n ≥ 3. N.S. indicates not significant. Scale bar: 200 μm (A–C’’, E–H’’). Genotypes: (A–A’’) btl-Gal4/+;P[B123]-RFP-moe/+; (B–B’’) btl-Gal4/+;P[B123]/UAS-ds; (C–C’’) btl-Gal4/UAS-ft;P[B123]/+; (E–E’’) btl-Gal4/+;P[B123]-RFP-moe/UAS-fat2RNAi; (F–F’’) btl-Gal4/+;P[B123]-RFP-moe/UAS-crbRNAi; (G–G’’) btl-Gal4/+;P[B123]-RFP-moe/UAS-yurtRNAi; (H–H’’) btl-Gal4/+;P[B123]-RFP-moe/UAS-scbRNAi. (J) btl-Gal4/+;P[B123]-RFP-moe/+ (control); btl-Gal4/+;P[B123]-RFP-moe/UAS-enaRNAi; btl-Gal4/UAS-fakRNAi;P[B123]-RFP-moe/+; btl-Gal4/+;P[B123]-RFP-moe/UAS-E-cadRNAi; btl-Gal4/+;P[B123]-RFP-moe/UAS-robo2RNAi.

Protrusions extending from the leading edge of progenitors.
The confocal image showing the filopodia at the leading edge of migrating tracheal progenitors in control (A) and upd2RNAi flies (B). Arrows denote anterior–posterior (A–P) axis. Dashed lines indicate dorsal trunk. (C) Bar graph depicting the number of filopodia extending from leading edge. Error bars represent SEM, n = 8. Expression of bnl-lacZ in the trachea of control (D) and stat92ERNAi (E) flies. Scale bar: 100 μm (A, B), 500 μm (D, E). Genotypes: (A) lsp2-Gal4,P[B123]-RFP-moe/+; (B) lsp2-Gal4,P[B123]-RFP-moe/UAS-upd2RNAi.

The dependence of migration directionality on Ft/Ds system.
(A) Representative traces plot the migration distance relative to the origin, n ≥ 10. (B) Rose plot depicting the direction of cell movement. (C) Representative traces showing the movement of individual dsRNAi cells relative to their origin. (D) Rose plot depicting the movement direction of dsRNAi cells. (E) Representative traces showing the movement of individual ftRNAi cells relative to their origin. (F) Rose plot depicting the movement direction of ftRNAi cells. (G) Scatter plots represent the ratio (d/D) of straight-line length displacement (d) relative to the length of the migration track (D) of individual cell.
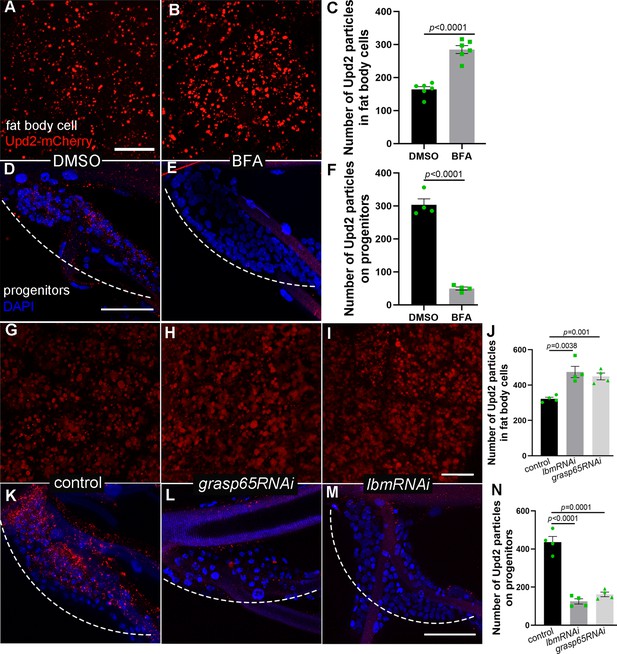
The production and transport of Upd2 from fat body.
The number of Upd2-mCherry-containing vesicles in fat body of control DMSO-fed (A) and BFA-treated L3 larvae (B). Larger view in lower magnification is provided in Figure 6—figure supplement 1. (C) Bar graph plots the number of Upd2-mCherry-containing vesicles in fat body. Error bars represent SEM, n = 6. The confocal image showing the number of Upd2-containing vesicles in progenitors of DMSO-fed control (D) and BFA-treated flies (E). (F) Bar graph plots the number of Upd2-mCherry-containing vesicles in progenitors. Error bars represent SEM, n = 4. (G) The number of Upd2-mCherry containing vesicles (red) in fat body. Upd2 accumulation in fat body was increased in the presence of grasp65RNAi (H) and lbmRNAi (I). (J) Bar graph plots the number of Upd2-mCherry-containing vesicles. Error bars represent SEM, n = 4. The Upd2 vesicles (red) in tracheal progenitors (DAPI) in control (K), grasp65RNAi (L), and lbmRNAi (M) flies. Dashed lines outline tracheal progenitors. (N) Bar graph plots the number of Upd2-mCherry-containing vesicles in progenitors. Error bars represent SEM, n = 4. Scale bar: 20 μm (A, B, G–I), 50 μm (D, E, K–M). Genotypes: (A–G, K) UAS-upd2-mCherry/+;lsp2-Gal4/+; (H, L) UAS-upd2-mCherry/UAS-grasp65RNAi;lsp2-Gal4/+; (I, M) UAS-upd2-mCherry/+;lsp2-Gal4/UAS-lbmRNAi.
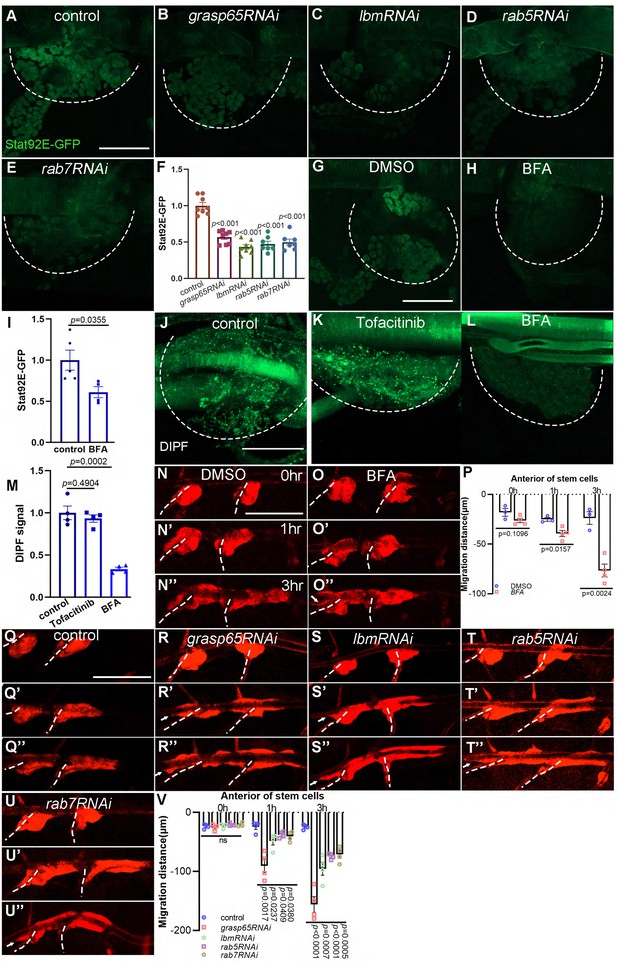
The dependence of tracheal progenitors on vesicle trafficking.
The expression of Stat92E-GFP in tracheal progenitors of control (A), grasp65RNAi (B), lbmRNAi (C), rab5RNAi (D), and rab7RNAi (E) flies. The progenitors are outlined by dashed lines. (F) Bar graph plots the relative expression of Stat92E-GFP. Error bars represent SEM, n = 7. The expression of Stat92E-GFP in tracheal progenitors in DMSO-fed (G) and BFA-treated (H) flies. (I) Bar graph plots the relative expression of Stat92E-GFP. Error bars represent SEM, n = 5. (J) The signal of DIPF reporter in tracheal progenitors. (K) The effects of Tofacinib (JAK inhibitor) on DIPF reporter in progenitors. (L) The effects of Brefeldin A on DIPF reporter in progenitors. Dashed lines outline tracheal progenitors. (M) Bar graphs showing the signal of DIPF reporter. Error bars represent SEM, n = 4. Migration of tracheal progenitors in DMSO-fed flies (N–N’’) and BFA-treated flies (O–O’’). (P) Bar graph plots migration distance of anterior movement. Error bars represent SEM, n ≥ 3. (Q–U’’) Migration of tracheal progenitors at 0 hr APF (Q), 1 hr APF (Q’), and 3 hr APF (Q’’). The confocal images showing the tracheal progenitors in control (Q–Q’’), grasp65RNAi (R–R’’), lbmRNAi (S–S’’), rab5RNAi (T–T’’), and rab7RNAi (U–U’’) flies. (V) Bar graph plots the migration distance of anterior movement. Error bars represent SEM, n = 4. Scale bar: 50 μm (A–E, G, H, J–L), 200 μm (N–O’’, Q–U’’). Genotypes: (A, G, H) lsp2-Gal4,Stat92E-GFP/+; (B) UAS-grasp65RNAi/+;lsp2-Gal4,Stat92E-GFP/+; (C) lsp2-Gal4,Stat92E-GFP/UAS-lbmRNAi; (D) UAS-rab5RNAi/+;lsp2-Gal4,Stat92E-GFP/+; (E) UAS-rab7RNAi/+;lsp2-Gal4,Stat92E-GFP/+; (J–L) btl-Gal4/UAS-DIPF; (N–Q’’) lsp2-Gal4,P[B123]-RFP-moe/+; (R–R’’) lsp2-Gal4,P[B123]-RFP-moe/UAS-grasp65RNAi; (S–S’’) lsp2-Gal4,P[B123]-RFP-moe/+;UAS-lbmRNAi/+; (T–T’’) UAS-rab5RNAi/+;lsp2-Gal4,P[B123]-RFP-moe/+; (U–U’’) UAS-rab7RNAi/+;lsp2-Gal4,P[B123]-RFP-moe/+.
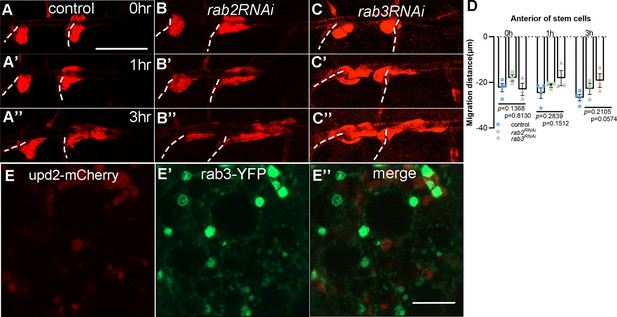
The roles of Rabs in transport of Upd2.
Migration of tracheal progenitors. The migration of progenitors at 0 hr APF, 1 hr APF, and 3 hr APF in control (A–A’’), rab2RNAi (B–B’’), rab3RNAi (C–C’’). (D) Bar graph plots the migration distance of anterior movement. Error bars represent SEM, n ≥ 3. (E–E’’) The confocal images showing the colocalization between Upd2 (red) (E) and Rab3 (GFP) (E’). (E’’) Merged images. Scale bar: 200 μm (A–C’’), 5 μm (E–E’’). Genotypes: (A–A’’) lsp2-Gal4,P[B123]-RFP-moe/+; (B–B’’) lsp2-Gal4,P[B123]-RFP-moe/UAS-rab2RNAi; (C–C’’) lsp2-Gal4,P[B123]-RFP-moe/UAS-rab3RNAi; (E–E’’) UAS-upd2-mCherry/UAS-rab3-YFP;lsp2-Gal4/+.

The roles of endocytic trafficking system in the transport of Upd2.
(A–D’’’) The confocal images showing the colocalization between Upd2 (red) (A–D) and Rab5 (GFP) (A’), Rab7 (GFP) (B’), Grasp65 (GFP) (C’), or Lbm (GFP). (A’’, B’’, C’’, D’’) Merged images. (D’’’) 3D high-magnification view of the boxed inset in D’’. (D’’’’) The Pearson’s correlation coefficient depicting colocalization between Lbm and Upd2 in fat body cells. The PLA (proximity ligation assay) assay showing the interaction between Upd2 and Rab5 (E, F), Rab7 (G, H), or Lbm (I, J). Co-immunoprecipitation assay showing physical interaction between Upd2 and Rab5 (K), Rab7 (L), or Lbm (M) in larval fat body. (N–T) The expression of Lbm-pHluorin in larval fat body and progenitors of control (N, O), rab5RNAi (P, Q), and rab7RNAi (R, S) flies. Dashed lines outline tracheal progenitors. Arrowheads point to Lbm-pHluorin puncta. DAPI signal indicates nuclei. (T) Schematic diagram depicting Upd2-operated disciplined migration of tracheal progenitors. Scale bar: 5 μm (A–D’’, E–J), 10 μm (N, P, R), 20 μm (O, Q, S). Genotypes: (A–A’’, E, F) UAS-upd2-mCherry/+;lsp2-Gal4/UAS-GFP-rab5; (B–B’’, G, H) UAS-upd2-mCherry/+;lsp2-Gal4/UAS-GFP-rab7; (C–C’’) UAS-upd2-mCherry/UAS-grasp65-GFP;lsp2-Gal4/+; (D–D’’’’, I–L) UAS-upd2-mCherry/UAS-lbm-GFP;lsp2-Gal4/+; (N, O) UAS-lbm-pHluorin/+;lsp2-Gal4/+; (P, Q) UAS-lbm-pHluorin/UAS-rab5RNAi;lsp2-Gal4/+; (R, S) UAS-lbm-pHluorin/UAS-rab7RNAi;lsp2-Gal4/+.
-
Figure 8—source data 1
Original files for western blot analysis displayed in Figure 8K–M.
- https://cdn.elifesciences.org/articles/100037/elife-100037-fig8-data1-v1.zip
-
Figure 8—source data 2
PDF file containing original western blots for Figure 8K–M, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100037/elife-100037-fig8-data2-v1.zip
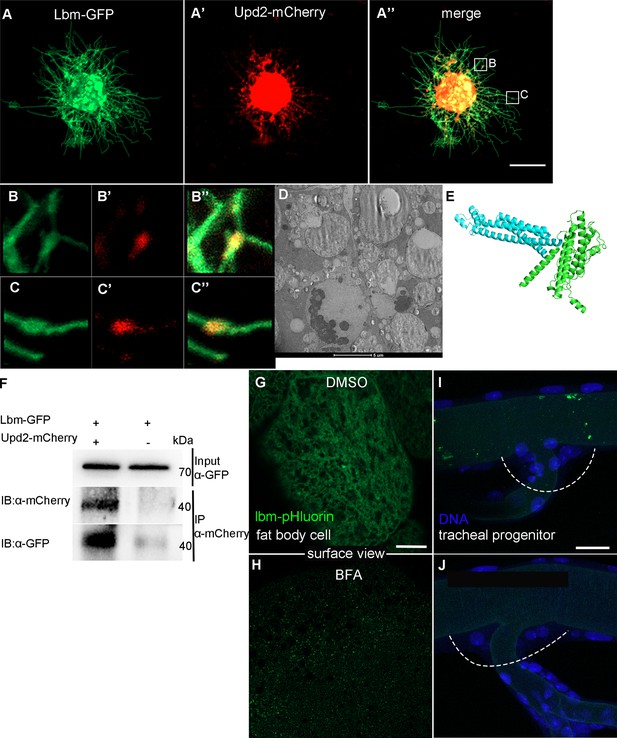
Vesicular transport of Upd2.
(A–C’’) Co-localization and interaction between Lbm and Upd2. Co-transfection of act-Gal4, UAS-lbm-GFP (A) and UAS-upd2-mCherry (A’) in S2 cells. (A’’) Merged image. (B–C’’) Confocal images showing that Lbm co-localizes with Upd2 in vesicles along the protrusions of S2 cell. (D) Electron microscopic image of Lbm-HRP vesicles (dark) in fat body. (E) A predicted interaction between Lbm (blue) and Upd2 (green). (F) Co-immunoprecipitation assay showing the interaction between Lbm and Upd2 in S2 cells. The expression of Lbm-pHluorin in control fat body (G) and BFA-treated flies (H). The expression of Lbm-pHluorin in control tracheal progenitors (I) and BFA-treated flies (J). The tracheal progenitors were outlined by dashed lines. Scale bar: 10 μm (A–A’’), 20 μm (G, H). Genotypes: (G–J) UAS-lbm-pHluorin/+;lsp2-Gal4/+.
-
Figure 8—figure supplement 1—source data 1
Original files for western blot analysis displayed in Figure 8—figure supplement 1F.
- https://cdn.elifesciences.org/articles/100037/elife-100037-fig8-figsupp1-data1-v1.zip
-
Figure 8—figure supplement 1—source data 2
PDF file containing original western blots for Figure 8—figure supplement 1F, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100037/elife-100037-fig8-figsupp1-data2-v1.zip
Videos
3D view of confocal image stack of tracheal progenitors and fat body.
Scale bar: 100 μm. Genotype: UAS-mCD8-GFP/+;lsp2-Gal4,P[B123]-RFP-moe/+.
The movement of tracheal progenitors in control and rpr.hid flies.
Scale bar: 100 μm. Genotypes: Gal80ts/+;lsp2-Gal4,P[B123]-RFP-moe/+ (control) and UAS-rpr-hid/+;Gal80ts/+;lsp2-Gal4,P[B123]-RFP-moe/+.
The movement of tracheal progenitors in control and upd2RNAi flies.
Scale bar: 100 μm. Genotypes: lsp2-Gal4,P[B123]-RFP-moe/+ (control) and lsp2-Gal4,P[B123]-RFP-moe/UAS-upd2RNAi.
The movement of tracheal progenitors in control and JAK/STAT pathway-perturbed flies.
Scale bar: 100 μm. Genotypes: btl-Gal4/+;P[B123]-RFP-moe/+ (control), btl-Gal4/+;P[B123]-RFP-moe/UAS-domeRNAi, btl-Gal4/+;P[B123]-RFP-moe/UAS-hopRNAi, and btl-Gal4/+;P[B123]-RFP-moe/UAS-stat92ERNAi.
The movement of tracheal progenitors in control and planar cell polarity (PCP) component aberrant flies.
Scale bar: 100 μm. Genotypes: btl-Gal4/+;P[B123]-RFP-moe/+ (control), btl-Gal4/UAS-dsRNAi;P[B123]-RFP-moe/+, and btl-Gal4/+;P[B123]-RFP-moe/UAS-ftRNAi.
The movement of tracheal progenitors in control and lbmRNAi flies.
Scale bar: 100 μm. Genotypes: lsp2-Gal4,P[B123]-RFP-moe/+ (control) and lsp2-Gal4,P[B123]-RFP-moe/+;UAS-lbmRNAi/+.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | upd2 | THFC | THU1331, THU1288 | |
| Gene (Drosophila melanogaster) | dome | THFC | THU0574, THU5825 | |
| Gene (Drosophila melanogaster) | hop | THFC | THU5762, TH201501042.S | |
| Gene (Drosophila melanogaster) | stat92E | THFC | THU0573, THU1915 | |
| Gene (Drosophila melanogaster) | lbm | THFC, BDSC | THU2602, BDSC27278 | |
| Gene (Drosophila melanogaster) | grasp65 | THFC | TH04282.N, THU1429 | |
| Gene (Drosophila melanogaster) | rab5 | THFC | TH02192.N, THU0671 | |
| Gene (Drosophila melanogaster) | rab7 | THFC | TH02539.N, THU2437 | |
| Gene (Drosophila melanogaster) | fj | THFC | THU201500988.S, THU1538 | |
| Gene (Drosophila melanogaster) | fat2 | THFC, VDRC | THU4120, VDRC27113 | |
| Gene (Drosophila melanogaster) | yurt | THFC, VDRC | THU1740, VDRC28674 | |
| Gene (Drosophila melanogaster) | crb | THFC | THU2783, THU5212 | |
| Gene (Drosophila melanogaster) | scb | THFC | THU3905, THU2707 | |
| Gene (Drosophila melanogaster) | ds | THFC, VDRC | THU2846, VDRC36219 | |
| Gene (Drosophila melanogaster) | ft | THFC, VDRC | TH201500989.S, VDRC9396 | |
| Genetic reagent (Drosophila melanogaster) | 10×Stat92E-GFP | THFC | THJ0273 | |
| Genetic reagent (Drosophila melanogaster) | Stat92E-GFP | BDSC | BDSC:38670 | |
| Genetic reagent (Drosophila melanogaster) | UAS-grasp65-GFP | BDSC | BDSC:8507 | |
| Genetic reagent (Drosophila melanogaster) | Ds::GFP | BDSC | BDSC:59425 | |
| Genetic reagent (Drosophila melanogaster) | UAS-GFP-Rab7 | BDSC | BDSC:9779 | |
| Genetic reagent (Drosophila melanogaster) | UAS-GFP-Rab5 | BDSC | BDSC:24616 | |
| Genetic reagent (Drosophila melanogaster) | hopTum/FM7C | BDSC | BDSC:8492 | |
| Genetic reagent (Drosophila melanogaster) | stat92EF | BDSC | BDSC:24757 | |
| Genetic reagent (Drosophila melanogaster) | domeG0264 | Kyoto | Kyoto:111866 | |
| Genetic reagent (Drosophila melanogaster) | Yurt::GFP | VDRC | VDRC:318067 | |
| Genetic reagent (Drosophila melanogaster) | Crb::GFP | VDRC | VDRC:318384 | |
| Genetic reagent (Drosophila melanogaster) | Ft::GFP | VDRC | VDRC:318477 | |
| Genetic reagent (Drosophila melanogaster) | Fj::GFP | VDRC | VDRC:318457 | |
| Genetic reagent (Drosophila melanogaster) | UAS-ft | This paper; Brittle et al., 2010 | ||
| Genetic reagent (Drosophila melanogaster) | UAS-ds | This paper; Brittle et al., 2012 | ||
| Genetic reagent (Drosophila melanogaster) | Fat2::GFP | This paper; Barlan et al., 2017 | ||
| Cell line (D. melanogaster) | S2 | CCTCC | GDC#0138 | Verified by DNA barcoding; without mycoplasma contamination |
| Cell line (Homo sapiens) | SKOV3 | ATCC | HTB-77 | Verified by STR genotyping; without mycoplasma contamination |
| Antibody | anti-GFP (Mouse monoclonal) | Abclonal | Cat# AE012 | WB (1:1000) |
| Antibody | anti-mCherry (Mouse polyclonal) | Abclonal | Cat# AE002 | WB (1:1000) |
| Antibody | Alexa Fluor 488 | Abclonal | Cat# AS053 | IF (1:200) |
| Antibody | Alexa Fluor 555 | Abclonal | Cat# AS007 | IF (1:200) |
| Antibody | Phalloidin Alexa Fluor 640 | Biotum | Cat# 00050 | IF (1:50) |
| Antibody | anti-tubulin (Rabbit polyclonal) | Baoke | Cat# BK7010 | WB (1:5000) |
| Antibody | anti-GFP (Rabbit polyclonal) | Invitrogen | Cat# A11122 | IF (1:400) |
| Antibody | HRP-conjugated Streptavidin | Proteintech | Cat# SA00001 | WB (1:5000) |
| Sequence-based reagent | α-tubulin84b _F | This paper | PCR primers | CACACCACCCTGGAGCATTC |
| Sequence-based reagent | α-tubulin84b _R | This paper | PCR primers | CCAATCAGACGGTTCAGGTTG |
| Sequence-based reagent | upd2_F | This paper | PCR primers | TCAATCCGTATCGCGGTCTG |
| Sequence-based reagent | upd2_R | This paper | PCR primers | AGAAGAGTCGCAGGTTGTGG |
| Sequence-based reagent | ds_F | This paper | PCR primers | ACAACCGAACTCGAACCGAA |
| Sequence-based reagent | ds_R | This paper | PCR primers | AGTAGCATCACACACAAGTGA |
| Sequence-based reagent | ft_F | This paper | PCR primers | CTGGATCGAGAGCAGCAGAG |
| Sequence-based reagent | ft_R | This paper | PCR primers | GACGGTAAATTCTCGCGCAC |
| Sequence-based reagent | fj_F | This paper | PCR primers | ATTACTCAAGCGGTTGGGGG |
| Sequence-based reagent | fj_R | This paper | PCR primers | CGGTTCCTGTTCCTGTCTCC |
| Sequence-based reagent | fat2_F | This paper | PCR primers | TATCTGCGCCCATACGCATT |
| Sequence-based reagent | fat2_R | This paper | PCR primers | TCTCATCGGCCTTGCTTTGT |
| Sequence-based reagent | yurt_F | This paper | PCR primers | GGTCAGCTCAGGGTGACTATC |
| Sequence-based reagent | yurt_R | This paper | PCR primers | ATTGGTAAGCTTGGCGTTGC |
| Sequence-based reagent | crb_F | This paper | PCR primers | CAGCAGTGTTTGAACGGTGG |
| Sequence-based reagent | crb_R | This paper | PCR primers | AGGCAGTGACCAATGGGG |
| Peptide, recombinant protein | Anti-FLAG M2 Magnetic Beads | Millipore | Cat# 8823 | |
| Commercial assay or kit | RNeasy Micro Kit | QIAGEN | Cat# 74004 | |
| Commercial assay or kit | SMART-Seq v4 Ultra low input RNA Kit | Takara | Cat# 634889 | |
| Commercial assay or kit | AMPure XP | Beckman Coulter | Cat# A63882 | |
| Commercial assay or kit | TruePrep DNA Library Prep Kit V2 | Vazyme | Cat# TD501 | |
| Chemical compound, drug | BXXP | APEXBIO | Cat# A8012 | |
| Software, algorithm | Fiji/ImageJ | NIH | RRID:SCR_002285 | |
| Software, algorithm | GraphPad Prism 8.0 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ | |
| Software, algorithm | Zen 3.1 | Zeiss | https://www.zeiss.com.cn/corporate/home.html | |
| Software, algorithm | PCA-flow | Bradski, G.79 | https://www.drdobbs.com/open-source/the-opencv-library | |
| Other | DAPI | VECTASHIELD | Cat# H1200 |