Inclusive, exclusive and hierarchical atlas of NFATc1+/PDGFR-α+ cells in dental and periodontal mesenchyme
Figures

Advanced genetic recombination and imaging techniques for craniofacial tissue analysis in transgenic models.
(A) Genetic recombination systems.The initial column lists various reporter alleles, while the subsequent second and third columns present the corresponding reporter constructs and the recombinases utilized. The final column provides the outcomes associated with the respective reporter alleles. Traditional reporter mechanisms comprise both single-color and multicolor systems (not shown in the figure), created through a singular recombination event. Advanced dual-reporter configurations encompass intersectional reporters, exclusive reporters, and nested reporters. (B) The craniofacial hard tissue, including maxilla and mandible of transgenic mice with multiple DNA recombinases-based genetic lineage tracing system, were obtained for advanced imaging technologies. (C–a) The tissue deep clearing procedure based on SUMIC, which needed to go through 4% PFA fixation, 0.5 M EDTA decalcification, different concentration of methanol dehydration, 5% H2O2 bleaching, ECI tissue clearing, and finally used the light-sheet system to directly image the whole tissue. (C–b) The traditional frozen section technology needs fixation, decalcification, dehydration and other steps, which takes about 2 weeks.

The observation of PDGFR-α+ and NFATc1+ cells in maxilla M1 of PdgfraCreER × Nfatc1DreER × LGRT mice (pulse) by whole-mount and high-speed imaging.
The whole-mount and high-speed imaging of (A) mouse molar with a tiling light-sheet microscope. (B) Schematic illustration of lineaging tracing in PdgfraCreER×Nfatc1DreER× LGRT mice. The mice were administrated with tamoxifen at D1 and D3, and sacrificed at D5. (C) The contoured M1 of maxilla, including pulp and PDL with virtual dentin shell (white) in buccal view (scale bar = 300 μm). (D) Image stack was displayed in buccal view, coronal view, and radicular view of pulp and PDL, respectively (scale bar = 300 μm). (E) An optical slice was acquired on the X-Z (scale bar = 300 μm) and X-Y direction (scale bar = 400 μm) to display the pulp and PDL.
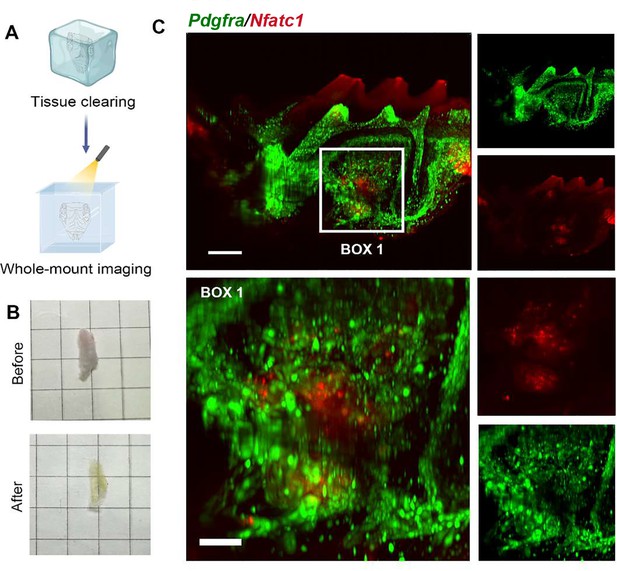
SUMIC procedure achieves whole-mount transparency of mice maxilla and enables imaging of dental, periodontal and alveolar bone.
(A) The tissue-clearing (TC) and whole-mount imaging procedure of mice maxilla. (B) The images before & after the TC procedure of mice maxilla. (C) The distribution of PDGFR-a+ and NFATc1+ cells from the section of XZ axis after 3D reconstruction of TC imaging. The image was from PdgfraCreER×Nfatc1DreER× LGRT mice (pulse). Box 1: alveolar bone. Scale bar = 300 μm for top row, 100 μm for bottom row.
Panoptic multicolor imaging of PDGFR-α+ cells(green) & NFATc1+ cells(red) in the pulp and PDL area of maxilla M1 of PdgfraCreER×Nfatc1DreER× LGRT mice (pulse), the whole-tissue imaging was achieved through TC procedure, related to Figure 2C.

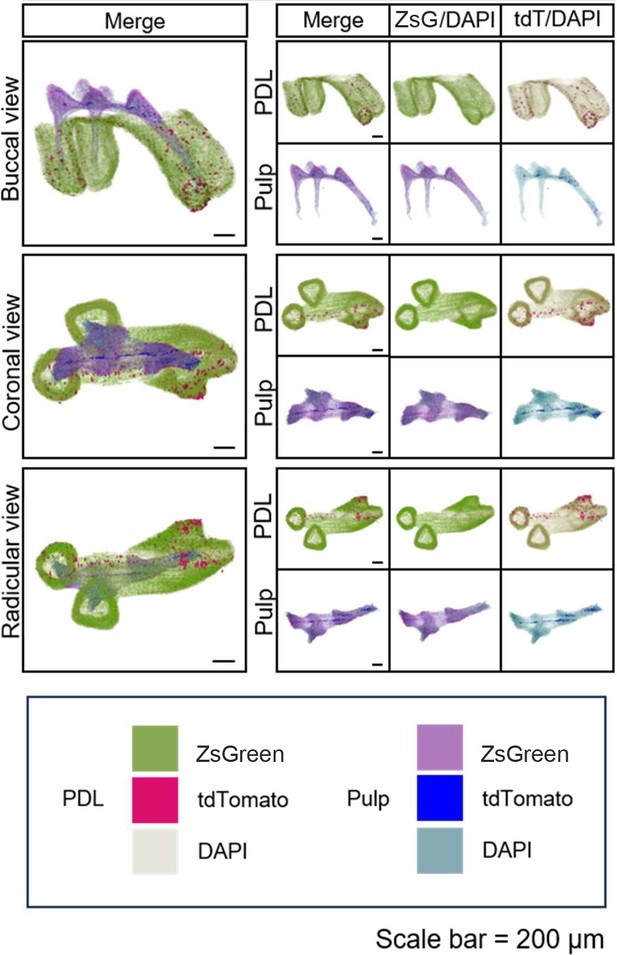
Using traditional serial section for imaging of maxilla of PdgfraCreER×Nfatc1DreER× LGRT mice (pulse).
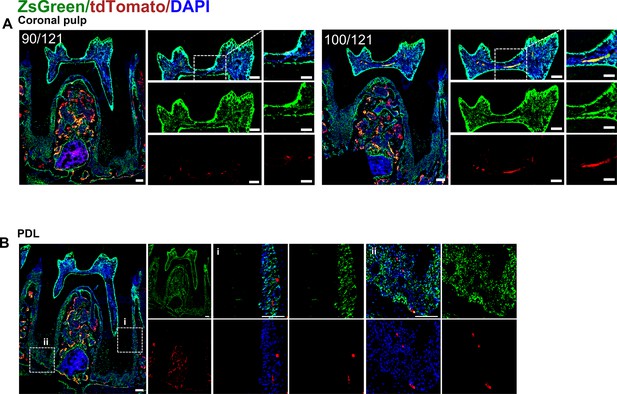
(A) Operation process of frozen section. Using this procedure, a total of 121 slices were collected in this sample. (B) Representative images acquired by confocal microscopy (scale bar = 200 μm). Box 1: coronal pulp, Box 2: root pulp, Box 3: PDL, scale bar = 100 μm. (C) Maxilla M1 after 3D reconstruction by imaris, including pulp and PDL with virtual dentin shell (white) in buccal view (scale bar = 300 μm). (D) Image stack was displayed in buccal view, coronal view, and radicular view of pulp and PDL, respectively (scale bar = 300 μm).
The total 121 slices of maxilla M1 of PdgfraCreER×Nfatc1DreER× LGRT mice sample (pulse).
The images were acquired by confocal microscope, ZsGreen+ cells in green, tdTomato+ cells in red, DAPI in blue.
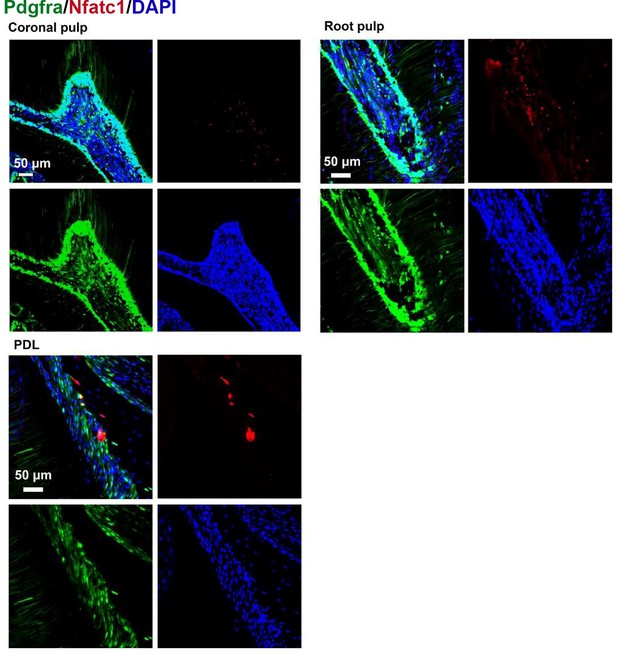
Representative images of coronal pulp, root pulp, and PDL of maxilla M1 of PdgfraCreER×Nfatc1DreER× LGRT mice sample (pulse).
The images were acquired by confocal microscope.
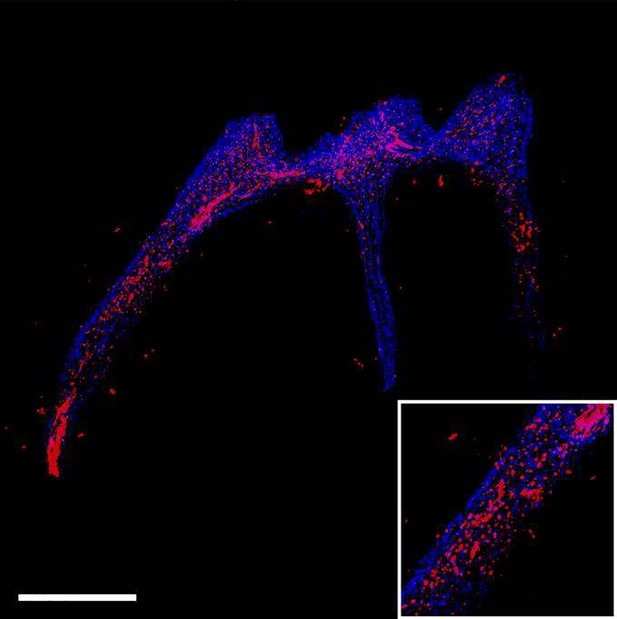
The tdTomato signal in pulp reconstructed by traditional serial section-based confocal imaging method (scale bar = 300 μm).
The sample was from PdgfraCreER×Nfatc1DreER× LGRT mice (pulse).
The discontinuities in the z-axis due to stratification of slices.
The image was reconstructed by serial sections of maxilla M1 of PdgfraCreER×Nfatc1DreER× LGRT mice sample (pulse).
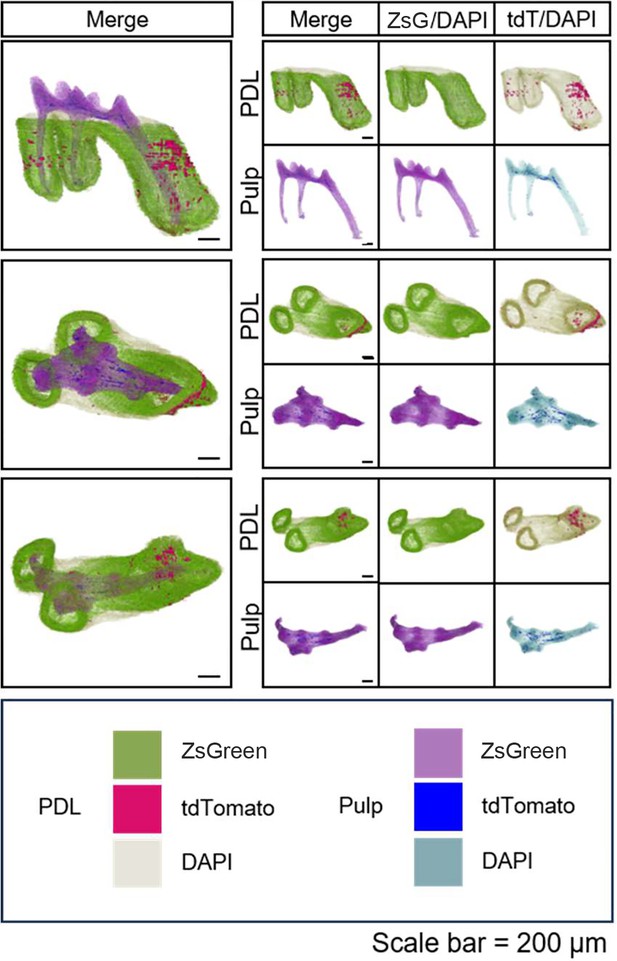
3D reconstruction of maxilla M1 of PdgfraCreER×Nfatc1DreER× LGRT mice (pulse) by DICOM-3D; in PDL: ZsGreen+ cells in green, tdTomato+ cells in rose red; in pulp: ZsGreen+ cells in purple, tdTomato+ cells in blue.
The image stack was also displayed in buccal view, coronal view, and radicular view of pulp and PDL, respectively.
Panoptic multicolor imaging of PDGFR-α+ cells & NFATc1+ cells in the pulp and PDL area of maxilla M1 of PdgfraCreER×Nfatc1DreER× LGRT mice (pulse), the whole-tissue imaging was reconstructed from serial sections, related to Figure 3C.
PDGFR-α+ cells in green, NFATc1+ cells in red and DAPI in blue.

Using traditional serial section for imaging of mandible of PdgfraCreER×Nfatc1DreER× LGRT mice (pulse).
(A) Schematic illustration of lineaging tracing in PdgfraCreER×Nfatc1DreER× LGRT mice. (B) Operation process of frozen section. Using this procedure, a total of 88 slices were collected in this sample. (C) Mandible M1 after 3D reconstruction by Imaris, including pulp and PDL with virtual dentin shell (white) in buccal view, coronal view and radicular view (scale bar = 200 μm). (D) 3D reconstruction of mandible M1 by DICOM-3D; in PDL, ZsGreen+ cells in green, tdTomato+ cells in rose red; in pulp, ZsGreen+ cells in purple, tdTomato+ cells in blue. The image stack was also displayed in buccal view, coronal view, and radicular view of pulp and PDL, respectively (D-b), scale bar = 200 μm. (D-a): The legend of (D-b).
The total 88 slices of mandible M1 of PdgfraCreER×Nfatc1DreER× LGRT mice sample (pulse).
The images were acquired by confocal microscope, ZsGreen+ cells in green, tdTomato+ cells in red, DAPI in blue.
Representative images of coronal pulp (A) and PDL (B) acquired by confocal microscope of mandible M1 of PdgfraCreER×Nfatc1DreER× LGRT mice sample (pulse).
The images were acquired by confocal microscope (Scale bar = 100 μm).
Panoptic multicolor imaging of PDGFR-α+ cells & NFATc1+ cells in the pulp and PDL area of mandible M1 of PdgfraCreER×Nfatc1DreER× LGRT mice (pulse), the whole-tissue imaging was reconstructed from serial sections, related to Figure 4C.
PDGFR-α+ cells in green, NFATc1+ cells in red and DAPI in blue.

The observation of lineage tracing of PDGFR-α+ and NFATc1+ cells in M1 by whole-mount and high-speed imaging.
(A) The flowchart of tracing. The mice were administrated with tamoxifen at D1 and D3, and sacrificed at D11. (B) The 3D images of contoured M1 of maxilla, including pulp and PDL with virtual dentin shell (white) in buccal view (scale bar = 300 μm). (D) Image stack was displayed in buccal view, coronal view, and radicular view of pulp and PDL, respectively. (E) An optical slice was acquired on the X-Z (scale bar = 400 μm) and X-Y direction to display the pulp and PDL (scale bar = 300 μm).
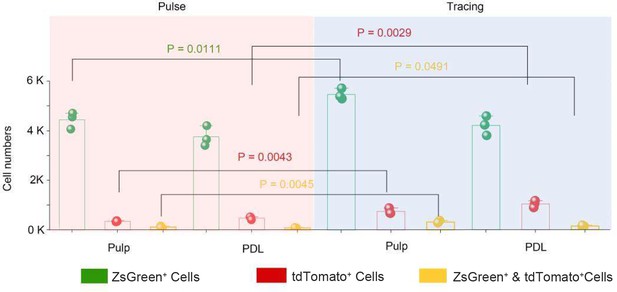
The number of PDGFR-α+ cells, NFATc1+ cells, and PDGFR-α+&NFATc1+ cells in pulp and PDL, respectively.
The quantification was done on a z stack of images in Imaris using the automatic spot detection feature.
-
Figure 5—figure supplement 1—source data 1
The tables present the source data for the number of PDGFR-α+ cells, NFATc1+ cells, and PDGFR-α+&NFATc1+ cells in pulp and PDL, respectively.
The quantification was done on a z stack of images in Imaris using the automatic spot detection feature.
- https://cdn.elifesciences.org/articles/100173/elife-100173-fig5-figsupp1-data1-v1.xlsx
Panoptic multicolor imaging of ZsGreen+ cells (green) & tdTomato+ cells (red) in the pulp and PDL area of maxilla M1 of PdgfraCreER×Nfatc1DreER× LGRT mice (tracing 11 days), the whole-tissue imaging was achieved through TC procedure, related to Figure 5B.

Using traditional serial section for imaging of maxilla of PdgfraCreER×Nfatc1DreER× LGRT mice (tracing 11 days).
(A) Using this procedure, a total of 117 slices were collected in this sample. (B) Representative images acquired by confocal microscopy (scale bar = 200 μm). Box 1: coronal pulp; Box 2: root pulp; Box 3: PDL (scale bar = 50 μm). (C) Maxilla M1 after 3D reconstruction by imaris, including pulp and PDL with virtual dentin shell (white) in buccal view (scale bar = 300 μm). (D) Image stack was displayed in buccal view, coronal view, and radicular view of pulp and PDL, respectively (scale bar = 300 μm).
The total 117 slices of maxilla M1 of PdgfraCreER×Nfatc1DreER× LGRT mice sample (tracing).
The images were acquired by confocal microscope, ZsGreen+ cells in green, tdTomato+ cells in red, DAPI in blue.
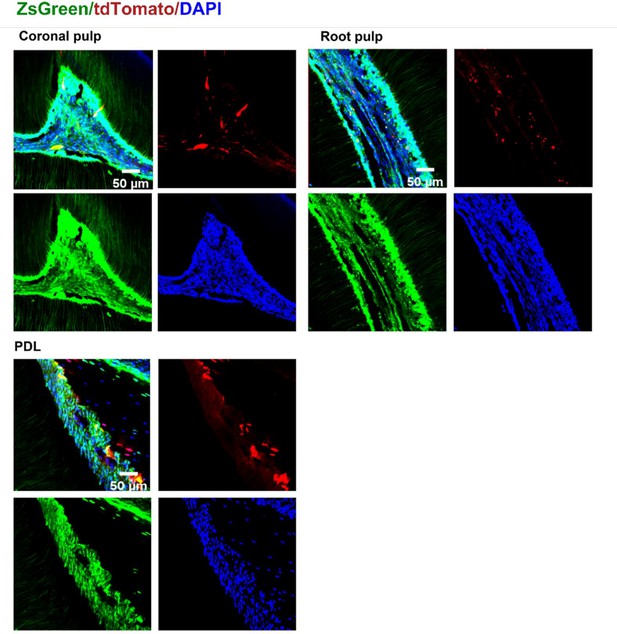
Representative images of coronal pulp, root pulp, and PDL of maxilla M1 of PdgfraCreER×Nfatc1DreER× LGRT mice sample (tracing).
The images were acquired by confocal microscope (scale bar = 50 μm).
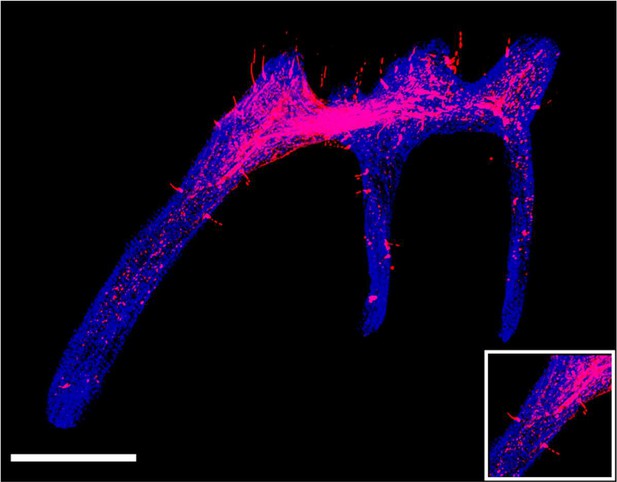
The tdTomato signal in pulp of maxilla M1 of PdgfraCreER×Nfatc1DreER× LGRT mice sample (tracing) reconstructed by traditional serial section-based confocal imaging method (scale bar = 300 μm).
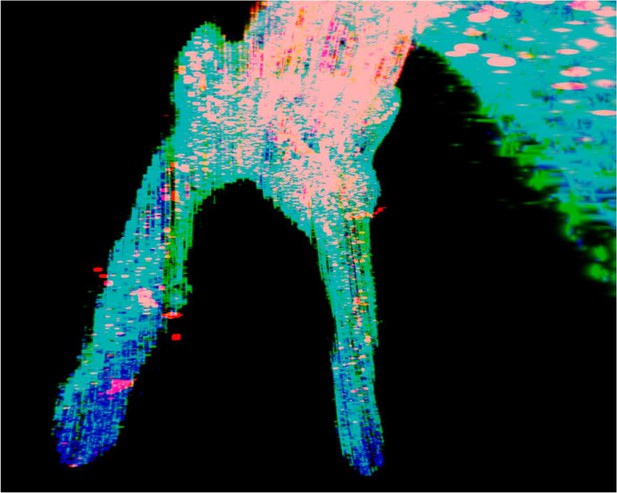
The discontinuities in the z-axis due to stratification of slices.
The image was reconstructed by serial sections of maxilla M1 of PdgfraCreER×Nfatc1DreER× LGRT mice sample (tracing) using Imaris. ZsGreen+ cells in green, tdTomato+ cells in red, DAPI in blue.
3D reconstruction of maxilla M1 of PdgfraCreER×Nfatc1DreER× LGRT mice (tracing) by DICOM-3D; PDL: ZsGreen+ cells in green, tdTomato+ cells in rose red; pulp: ZsGreen+ cells in purple, tdTomato+ cells in blue.
The image stack was also displayed in buccal view, coronal view, and radicular view of pulp and PDL, respectively.
Panoptic multicolor imaging of ZsGreen+ cells & tdTomato+ cells in the pulp and PDL area of maxilla M1 of PdgfraCreER×Nfatc1DreER× LGRT mice (tracing 11 days), the whole-tissue imaging was reconstructed from serial sections, related to Figure 6C.
ZsGreen+ cells in green, tdTomato+ cells in red and DAPI in blue.

MCIR tracing distinct cell populations simultaneously in the pulp and PDL.
(A) Schematic illustration of pulse in PdgfraCreER×Nfatc1DreER× LGRT mice. (B) Operation process of frozen section. Using this procedure, a total of 120 slices were collected in this sample. (C) 3D reconstruction of mandible M1 using Imaris, including pulp and PDL with virtual dentin shell (white) (scale bar = 200 μm). The image stack was displayed in buccal view, coronal view, and radicular view of pulp and PDL, respectively. (D) 3D reconstruction of mandible M1 using DICOM-3D, in which the distribution of NFATc1+ cells in pulp and PDL tissues can be more precisely perceived (D-b), scale bar = 200 μm. (D-a): The legend of (D-b).
All consecutive slices (a total of 120 slices) for imaging of mandible M1 of PdgfraCreER×Nfatc1DreER× LGRT mice (tracing 11 days).
The images were acquired by confocal microscope, ZsGreen+ cells in green, tdTomato+ cells in red, DAPI in blue.
Representative images of coronal pulp (A) and PDL (B) acquired by confocal microscope of mandible M1 of PdgfraCreER×Nfatc1DreER× LGRT mice sample (tracing).
Scale bar = 100 μm.
Panoptic multicolor imaging of ZsGreen+ cells (green) & tdTomato+ cells (red) in the pulp and PDL area mandible M1 of PdgfraCreER×Nfatc1DreER× LGRT mice (tracing 11 days), the whole-tissue imaging was reconstructed from serial sections, related to Figure 7C.
ZsGreen+ cells in green, tdTomato+ cells in red and DAPI in blue.

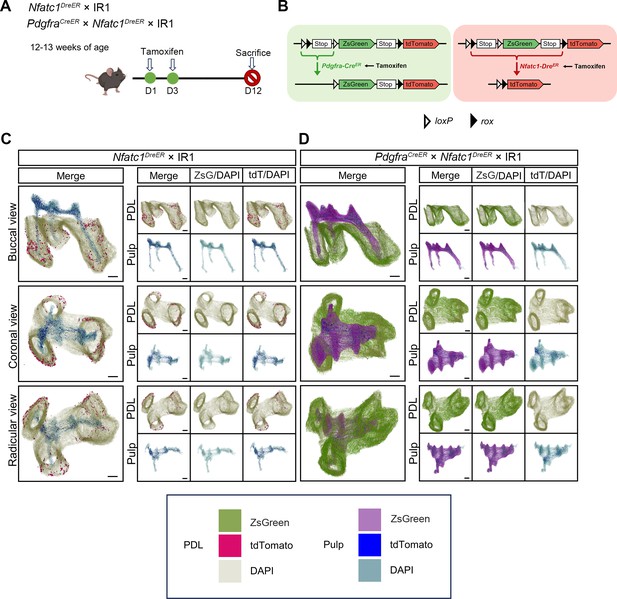
IR1 tracing distinct cell populations simultaneously in the pulp and PDL.
(A) Schematic illustration of lineaging tracing in PdgfraCreER× IR1, Nfatc1DreER× IR1 and PdgfraCreER×Nfatc1DreER× IR1 mice. (B) Schematic diagram of the IR1 working principle. (C) 3D reconstruction of maxilla M1 using Imaris, including pulp and PDL with virtual dentin shell (white) (scale bar = 200 μm). The image stack was displayed in buccal view, coronal view, and radicular view of PDL (D) and pulp (E), respectively.
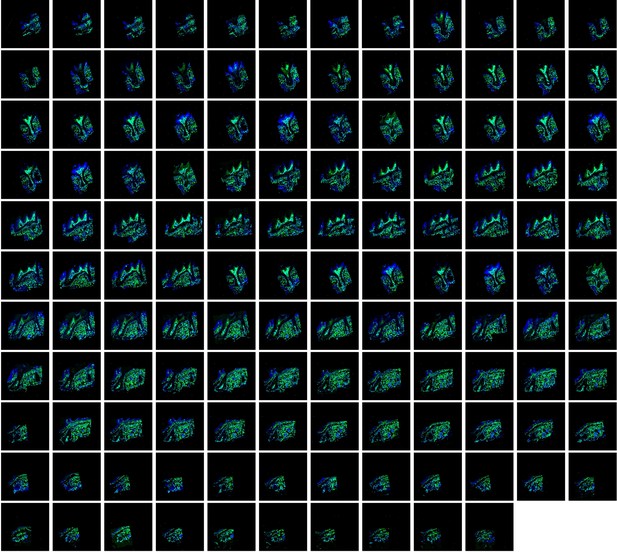
All consecutive slices (a total of 130 slices) for imaging of maxilla of PdgfraCreER × IR1 mice.
The images were acquired by confocal microscope, ZsGreen+ cells in green and DAPI in blue.
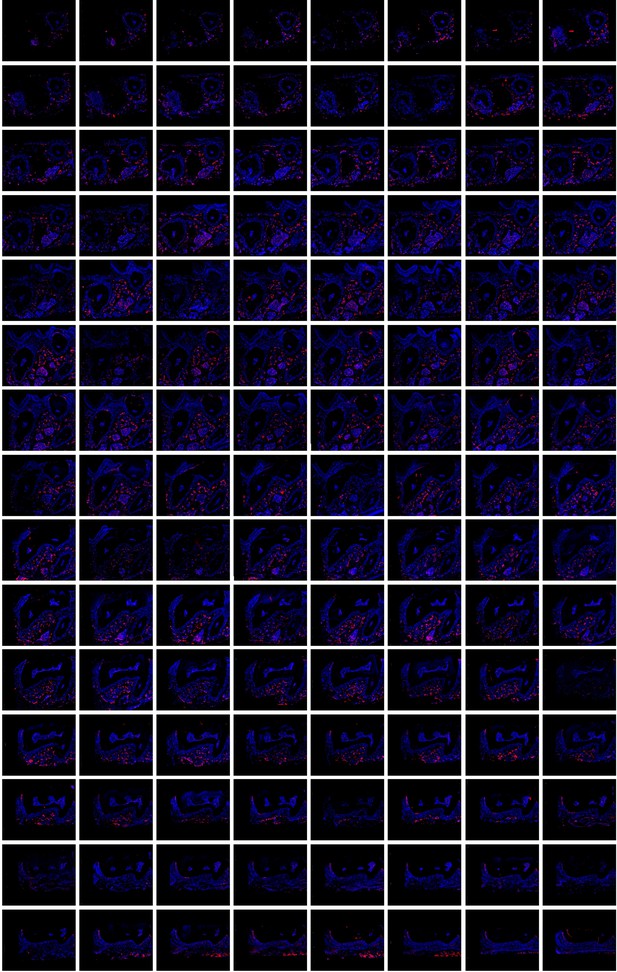
All consecutive slices (a total of 121 slices) for imaging of maxilla of Nfatc1DreER× IR1 mice.
The images were acquired by confocal microscope, tdTomato+ cells in red and DAPI in blue.
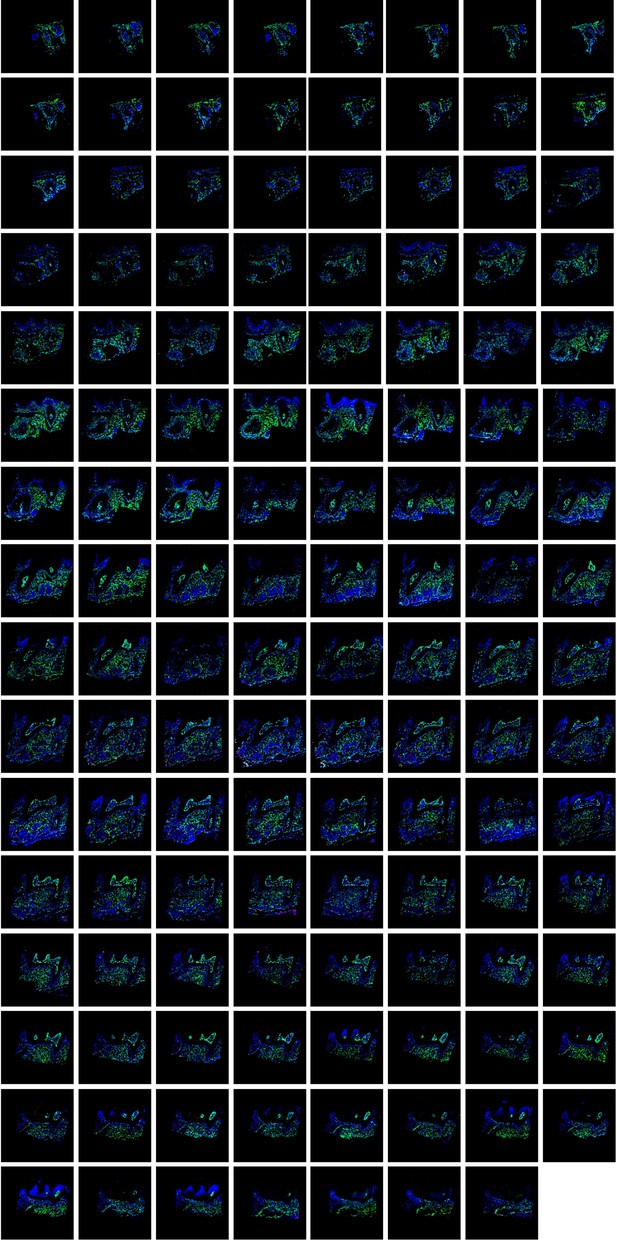
All consecutive slices (a total of 127 slices) for imaging of maxilla of PdgfraCreER ×Nfatc1DreER× IR1 mice sample.
The images were acquired by confocal microscope, ZsGreen+ cells in green, tdTomato+ cells in red, DAPI in blue.
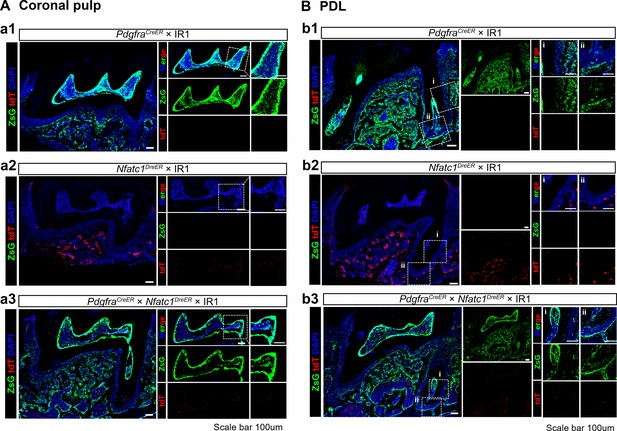
Representative images of coronal pulp (A) and PDL (B) acquired by confocal microscope of mandible M1 of PdgfraCreER × IR1 (a1, b1), Nfatc1DreER× IR1 (a2, b2) and PdgfraCreER×Nfatc1DreER× IR1 (a3, b3) mice.
Scale bar = 100 μm.
3D reconstruction of maxilla M1 by DICOM-3D of maxilla M1 of Nfatc1DreER× IR1 and PdgfraCreER ×Nfatc1DreER× IR1 mice.
(A) Schematic illustration of lineaging tracing in Nfatc1DreER× IR1 and PdgfraCreER×Nfatc1DreER× IR1 mice. (B) Schematic diagram of the IR1 working principle. (C-D) 3D reconstruction of maxilla M1 using DICOM-3D in Nfatc1DreER× IR1 (C) and PdgfraCreER×Nfatc1DreER× IR1 (D) mice, including pulp and PDL (scale bar = 200 μm). The image stack was displayed in buccal view, coronal view, andradicular view of PDL and pulp , respectively. In PDL, ZsGreen+ cells in green, tdTomato+ cells in rose red; in pulp, ZsGreen+ cells in purple, tdTomato+ cells in blue.
The image stack was displayed in buccal view, coronal view, and radicular view. Scale bar: 200 μm.
Panoptic multicolor imaging of ZsGreen+ cells in the pulp and PDL area of maxilla M1 of PdgfraCreER× IR1 mice, the whole-tissue imaging was reconstructed from serial sections, related to Figure 8.
ZsGreen+ cells in green and DAPI in blue.
Panoptic multicolor imaging of tdTomato+ cells in the pulp and PDL area of maxilla M1 of NFATc1DreER× IR1 mice, the whole-tissue imaging was reconstructed from serial sections, related to Figure 8.
tdTomato+ cells in red and DAPI in blue.
Panoptic multicolor imaging of ZsGreen+ cells and tdTomato+ cells in the pulp and PDL area of maxilla M1 of PdgfraCreER×NFATc1DreER× IR1 mice, the whole-tissue imaging was reconstructed from serial sections, related to Figure 8.
ZsGreen+ cells in green, tdTomato+ cells in red and DAPI in blue.

Ablation of PDGFR-α+ Cells Disrupts the morphology of dental pulp and periodontal ligament tissues.
(A) Schematics of tamoxifen induction. (B) Schematic diagram of the DTA working principle. (C, E) Representative H&E images of pulp (C) and PDL (E) of mandible M1 in PdgfraCreER ×DTA and control mice. (D, F) Masson trichrome staining of pulp (D) and PDL (F) of mandible M1 in PdgfraCreER ×DTA and control mice. Arrows in (C, D) indicate the odontoblast cell layer. Dotted lines in (E, F) outline ROI of the PDL. D: dentin; AB: alveolar bone. Scale bar: 50 μm.
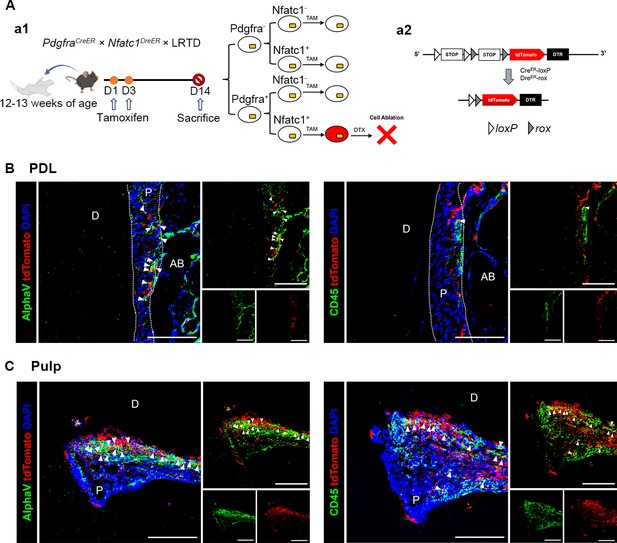
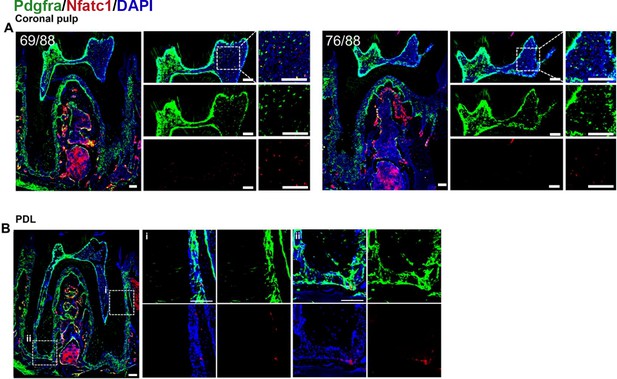
The identification of the types of PDGFR-α+ and NFATc1+ cells (red) of dental pulp and periodontal ligament tissues.
(A) a1: Schematic illustration of lineaging tracing in PdgfraCreER×Nfatc1DreER× LRTD mice. a2: The mice were administrated with tamoxifen at D1 and D3, and sacrificed at D14. (B, C) Representative IF images of PDL (B) and pulp (C) of mandible M1 in PdgfraCreER×Nfatc1DreER× LRTD mice showing the MSCs marker AlphaV (left) and hematopoietic marker CD45 (right). Arrows in (B, C) indicate the co-localization of AlphaV/CD45 and tdTomato. D: dentin; AB: alveolar bone. P: PDL (B), pulp (C). Scale bar: 100 μm.
Videos
Panoptic multicolor imaging of PDGFR-α+ cells (green) & NFATc1+ cells (red) in cranium of PdgfraCreER×Nfatc1DreER× LGRT mice (pulse), the whole-tissue imaging was achieved through TC procedure.
Panoptic multicolor imaging of PDGFR-α+ cells (green) and NFATc1+ cells (red) in cranial sagittal suture of PdgfraCreER×Nfatc1DreER× LGRT mice (pulse), the whole-tissue imaging was achieved through TC procedure.
Panoptic multicolor imaging of PDGFR-α+ cells (green) and NFATc1+ cells (red) in cranial coronal suture of PdgfraCreER×Nfatc1DreER× LGRT mice (pulse), the whole-tissue imaging was achieved through TC procedure.
Tables
| Methods | Durations | References |
|---|---|---|
| MarShie tissue clearing procedure | 13-15 days | Nat Commun. 2024, 15, 1764 |
| FDISCO clearing procedure | 4.5 days | Sci Adv. 2019, 5, eaau8355. |
| PEGASOS method | 12 days | Cell Research. 2018, 28, 803-818. |
| iDISCO | 4.5 days | J Comp Neurol. 2024 532, e25582. |





