Science Forum: Antibody characterization is critical to enhance reproducibility in biomedical research
Figures
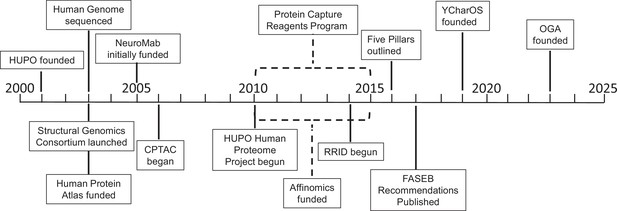
Key projects related to antibody characterization.
Timeline showing when the projects discussed in this article were started: the Protein Capture Reagents Program and the Affinomics project were funded for five years (as indicated by dashed lines). CPTAC: Clinical Proteomic Tumor Analysis Consortium; FASEB: Federation of American Societies of Experimental Biology; HUPO: Human Proteome Organization; OGA: Only Good Antibodies; RRID: Research Resource Identifier; YCharOS: Antibody Characterization through Open Science.
Tables
The ‘five pillars’ of antibody characterization.
In 2016 an ad hoc International Working Group for Antibody Validation introduced the five pillars of antibody validation/characterization: (i) genetic strategies; (ii) orthogonal strategies; (iii) (multiple) independent antibody strategies; (iv) recombinant strategies (originally called “expression of tagged proteins”); (v) capture MS strategies (Uhlen et al., 2016). In this table each pillar/strategy (left column) is followed by a brief description of the pillar/strategy, an indication of specificity, example applications for use, and pitfalls. Adapted from Waldron, 2022 and used with permission.
| Pillar/strategy | Description | Specificity | Example applications | Pitfalls | |
|---|---|---|---|---|---|
| i | Genetic strategies | Knock-out/ knock-down target gene | High | WB, IHC, IF, ELISA, IP | Requires a genetically tractable system and awareness of potential confounders (such as alternative isoforms) |
| ii | Orthogonal strategies | Compare results from Ab-dependent and Ab-independent experiments | Varies | WB, IHC, IF, ELISA | Requires variable expression of the target and cannot entirely rule out non-specific binding to similar proteins |
| iii | Independent antibody strategies | Compare results from experiments using unique Abs to the same target | Medium | WB, IHC, IF, ELISA, IP | Requires the purchase of multiple Abs and knowledge of their epitopes |
| iv | Recombinant strategies | Experimentally increase target protein expression | Medium | WB, IHC, IF | Overexpression of exogenous protein can lead to overconfidence in the specificity of the Ab |
| v | Capture MS strategies | Use MS to identify protein captured by Ab | Low | IP | Requires access to MS and it can be challenging to distinguish between Ab binding target vs protein bound to target |
-
Ab: antibody; ELISA: enzyme-linked immunosorbent assay; IF: immunofluorescence; IHC: immunohistochemistry; IP: immunoprecipitation; MS: mass spectrometry; WB: Western blotting.





