Magnesium modulates phospholipid metabolism to promote bacterial phenotypic resistance to antibiotics
Figures
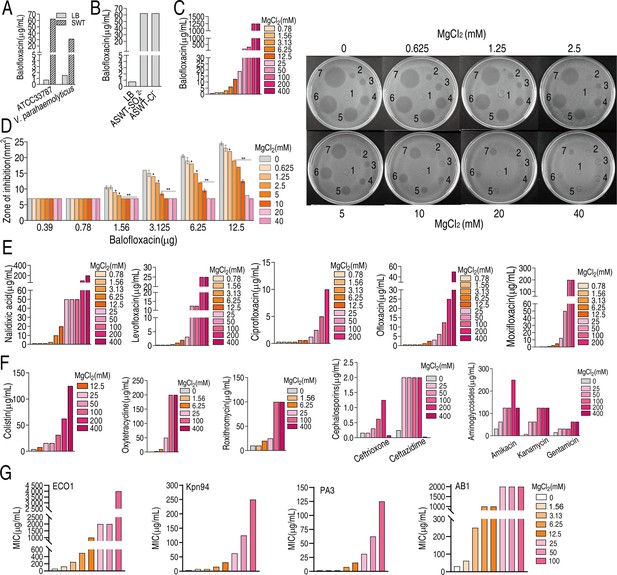
Magnesium promotes bacterial resistance to antibiotics.
(A) Minimal inhibitory concentration (MIC) of ATCC33787 and V. parahaemolyticus to balofloxacin (BLFX) in artificial seawater (ASWT) or Luria–Bertani (LB) medium as determined using the microtiter-dilution method. (B) MIC of ATCC33787 to BLFX in ASWT with additional MgSO4 or MgCl2 as determined using the microtiter-dilution method. (C) MIC of ATCC33787 to BLFX in ASWT at the indicated concentrations of MgCl2 as determined using the microtiter-dilution method. (D) MIC of ATCC33787 to BLFX at the indicated concentrations of BLFX and MgCl2 as determined by the Oxford cup test. The numbers 1, 2, 3, 4, 5, 6, and 7 represent 0, 0.39, 0.78, 1.56, 3.125, 6.25, and 12.5 µg BLFX, respectively. (E) MIC of ATCC33787 to other quinolones in ASWT at the indicated concentrations of MgCl2 as determined using the microtiter-dilution method. (F) MIC of ATCC 33787 to other classes of antibiotics in ASWT at the indicated concentrations of MgCl2 as determined using the microtiter-dilution method. (G) MIC of carbapenem-resistant Escherichia coli, carbapenem-resistant Klebsiella pneumoniae, carbapenem-resistant Pseudomonas aeruginosa, and carbapenem-resistant Acinetobacter baumannii isolates to BFLX at the indicated concentrations of MgCl2.
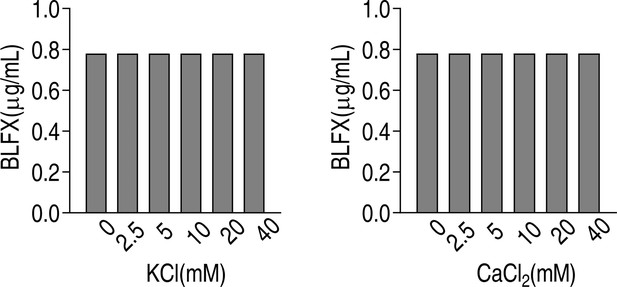
Minimal inhibitory concentration (MIC) of ATCC33787 to balofloxacin (BLFX) in the absence or presence of the indicated concentrations of KCl or CaCl2 in M9 media.
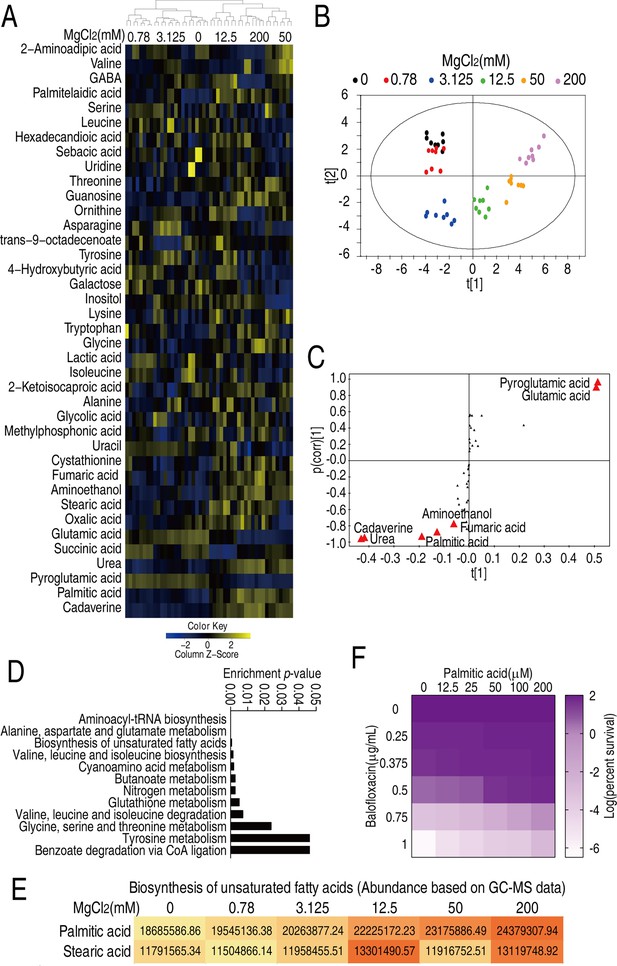
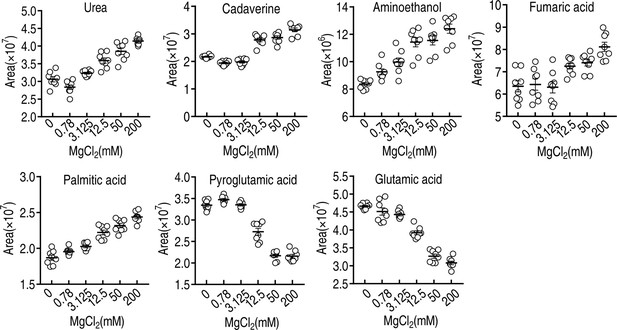
Mg2+-induced metabolomic change.
(A) Differential metabolomes in the absence or presence of the indicated concentrations of MgCl2. The yellow and blue colors indicate an increase or decrease in metabolite levels relative to the median metabolite level, respectively (see color scale). Euclidean distance calculations were used to generate a heatmap that shows clustering of the biological and technical replicates of each treatment. (B) Orthogonal partial least square discriminant analysis (OPLS-DA) of different MgCl2-induced metabolome concentrations. Each dot represents a technical replicate of samples in the plot. (C) S-plot generated from OPLS-DA. Predictive component p[1] and correlation p(corr)[1] differentiate 0, 0.78, and 3.125 mM MgCl2 from 12.5, 50, and 200 mM MgCl2. Predictive component p[2] and correlation p(corr)[2] separate 0, 0.78, 50, and 200 mM MgCl2 from 3.125 and 12.5 mM MgCl2. The triangle represents metabolites in which candidate biomarkers are marked. (D) Enriched pathways by differential abundances of metabolites. (E) Areas of the peaks of palmitic acid and stearic acid generated by GC-MS analysis. (F) Synergy analysis for balofloxacin (BLFX) with palmitic acid for V. alginolyticus. Synergy was performed by comparing the dose needed for 50% inhibition of the synergistic agents (white) and non-synergistic (i.e., additive) agents (purple).
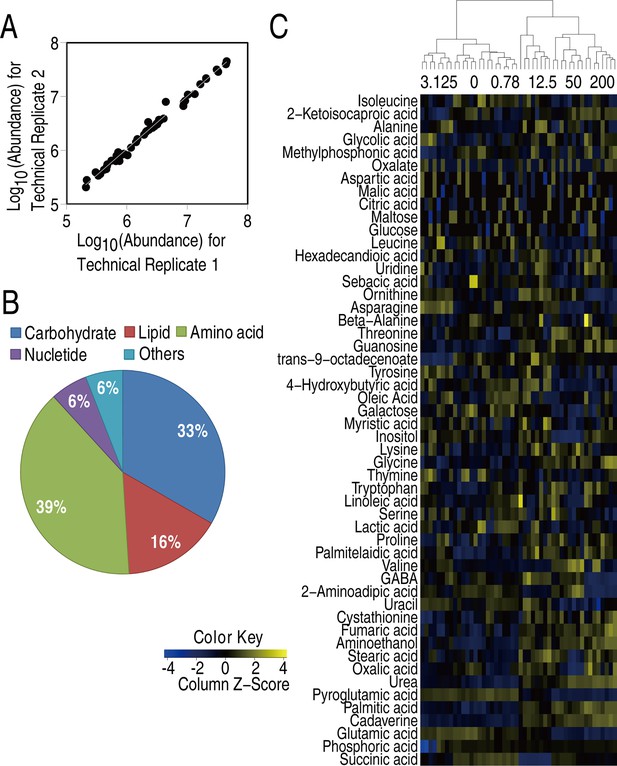
Metabolic profiles of V. alginolyticus in different concentrations of MgCl2.
(A) Reproducibility. (B) Percentage of metabolites in every category. (C) Heatmap of metabolites.
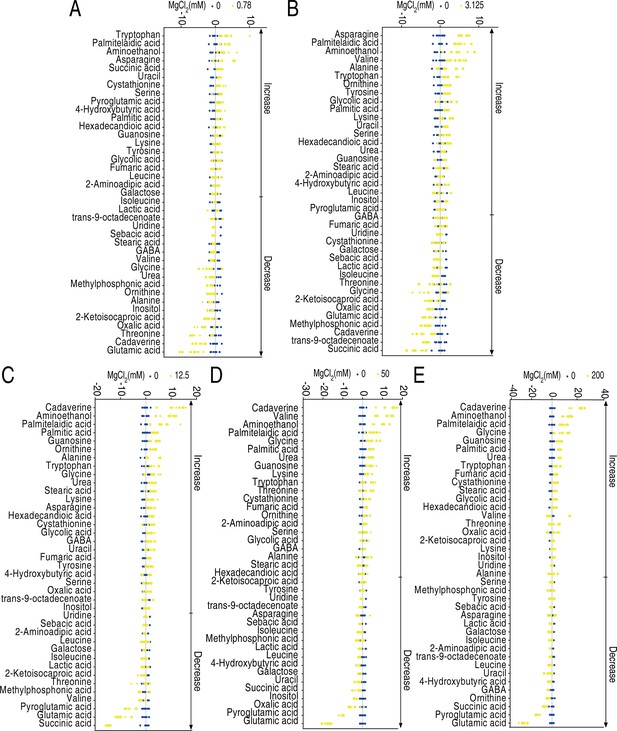
Heatmap and Z score plots of differential metabolites.
(A) Heatmap of differential metabolites. (B–F) Z score plots of differential metabolites.
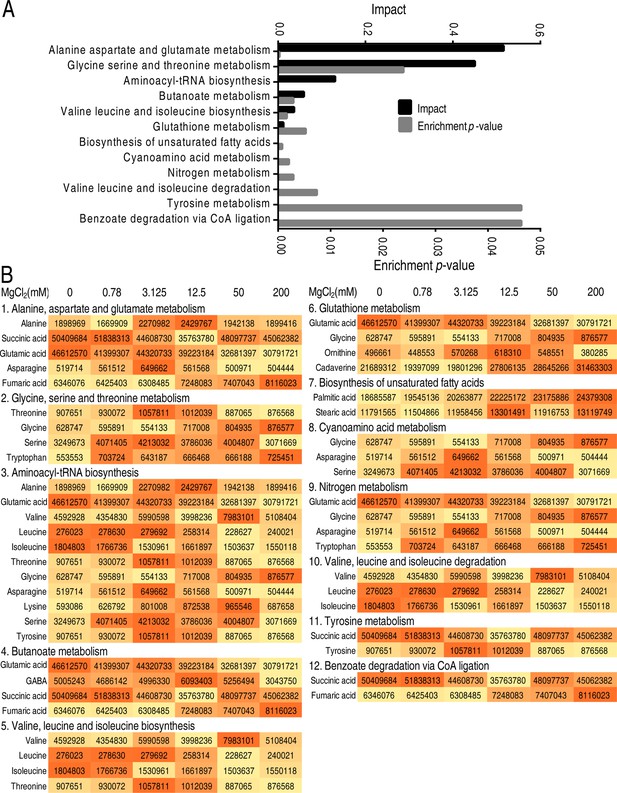
Pathway enrichment of differential metabolites.
(A) Pathway enrichment of differential metabolites. (B) Differential metabolites in enriched pathways.
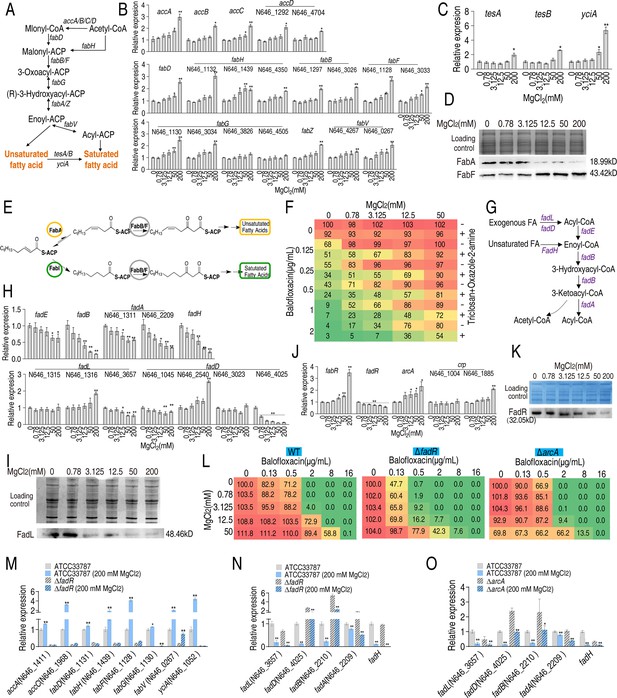
Mg2+ promotes fatty acid biosynthesis.
(A) Diagram of fatty acid biosynthesis. (B) qRT-PCR for expression of fatty acid biosynthesis genes in the absence or presence of indicated concentrations of MgCl2 (n = 4). (C) qRT-PCR for expression of genes involved in converting unsaturated fatty acids to saturated fatty acids in the absence or presence of indicated MgCl2 levels (n = 4). (D) Western blot for the abundance of proteins responsible for converting unsaturated fatty acids to saturated fatty acids in the absence or presence of indicated concentrations of MgCl2. (E) Diagram of saturated and unsaturated acid biosynthesis. (F) Synergy analysis of balofloxacin with triclosan + oxazole-2-amine for ATCC33787 (n = 3). The percentage of bacterial survival was quantified in the presence of indicated MgCl2 levels and/or balofloxacin plus triclosan and oxazole-2-amine and used to construct the isobologram. Synergy is represented using a color scale or an isobologram, which compares the dose needed for 50% inhibition of synergistic agents (blue) and non-synergistic (i.e., additive) agents (red). (G) Diagram of fatty acid degradation. (H) qRT-PCR for the expression of genes encoding fatty acid degradation in the absence or presence of the indicated concentrations of MgCl2 (n = 4). (I) Western blot for the abundance of FadL in the absence or presence of the indicated concentrations of MgCl2. (J) qRT-PCR for the expression of fatty acid biosynthesis regulatory genes in the absence or presence of the indicated concentrations of MgCl2 (n = 4). (K) Western blot for the abundance of FadR in the absence or presence of the indicated concentrations of MgCl2. (L) Synergy analysis for MgCl2 with BLFX for ATCC33787 (WT), ΔfadR, and ΔarcA. Synergy is represented using a color scale or an isobologram, which compares the dose needed for 50% inhibition of the synergistic agents (blue) and non-synergistic (i.e., additive) agents (red) (n = 3). (M–NO) qRT-PCR for expression of genes involved in fatty acid biosynthesis (M) and degradation (N, O) in ATCC33787, ΔfadR or/and ΔarcA (O) in the presence or absence of 200 mM MgCl2. Whole-cell lysates resolved by SDS-PAGE gel were stained with Coomassie brilliant blue as loading control (D), (I), and (K). Results are displayed as means ± SD, and statistically significant differences are identified by Kruskal–Wallis followed by Dunn’s multiple comparison post hoc test unless otherwise indicated. *p<0.05 and **p<0.01.
-
Figure 3—source data 1
File containing original western blots in Figure 3, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100427/elife-100427-fig3-data1-v1.zip
-
Figure 3—source data 2
Original files for western blot analysis displayed in Figure 3.
- https://cdn.elifesciences.org/articles/100427/elife-100427-fig3-data2-v1.zip
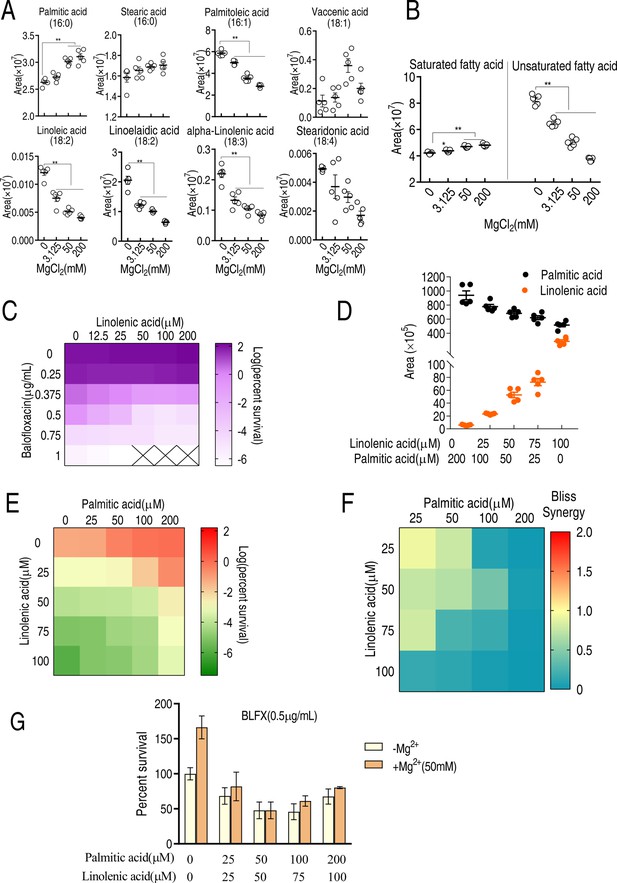
LC-MS targeted 16-carbon and 18-carbon fatty acids and the role of palmitic acids and linolenic acids in balofloxacin (BLFX) resistance.
(A) Scatter plots of 16-carbon and 18-carbon fatty acids, detected by LC-MS. The area indicates the area of the peak of the metabolite in total ion chromatography using GC-MS. (B) Scatter plots of total saturated fatty acids and unsaturated fatty acids with 16-carbon and 18-carbon (A). (C) Synergy analysis for BLFX with linolenic acid for ATCC 33787. Synergy is represented using a color scale or an isobologram, which compares the dose needed for 50% inhibition of synergistic agents (while) and non-synergistic (i.e., additive) agents (purple) (n = 3). (D) LC-MS for the abundance of intracellular linolenic acid and palmitic acid of ATCC33787 in synergy with the indicated exogenous linolenic acid and palmitic acid. (E) Synergy analysis of linolenic acid and palmitic acid in BLFX-mediated killing to ATCC33787. Synergy is represented using a color scale or an isobologram, which compares the dose needed for 50% inhibition of synergistic agents (blue) and non-synergistic (i.e., additive) agents (red). (F) Bliss analysis (E) (n = 3). (G) Percent survival of ATCC33787 in the presence of linolenic acid, palmitic acid, and BLFX with or without 50 mM MgCl2 (n = 3). Results are displayed as means ± SD, and statistically significant differences are identified by Kruskal–Wallis followed by Dunn’s multiple comparison post hoc test unless otherwise indicated. *p<0.05 and **p<0.01.
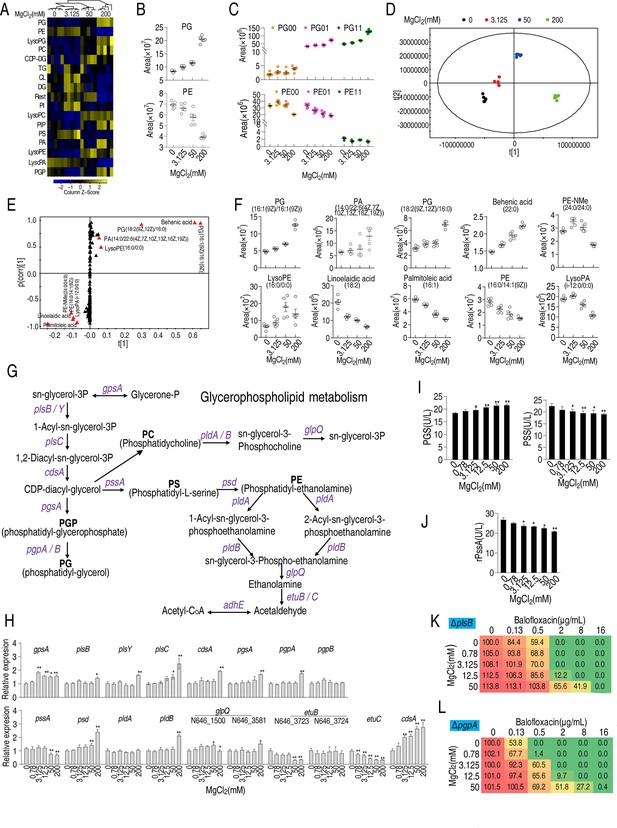
Effect of Mg2+ on phospholipid biosynthesis.
(A) Heatmap showing changes in differential lipid levels at the indicated concentration of MgCl2. (B) Abundance of ATCC33787 phosphatidylglycerol (PG) and phosphatidylethanolamine (PE) at the indicated concentrations of MgCl2. (C) Scatter plots of PG and PE at different saturation levels in the presence of the indicated MgCl2 levels. (D) Principal component analysis of different concentrations of MgCl2-induced phospholipids metabolomes. Each dot represents a technical replicate of samples in the plot. (E) S-plot generated from orthogonal partial least square discriminant analysis (OPLS-DA). Predictive component p[1] and correlation p (corr)[1] differentiated 0 and 3.125 mM MgCl2 from 50 and 200 mM MgCl2. (F) Scatter plot of biomarkers in data (E). (G) Diagram showing glycerophospholipid metabolism. (H) qRT-PCR of the expression of genes encoding glycerophospholipid metabolism in the absence of or at the indicated concentrations of MgCl2 (n = 4). (I) Phosphatidylglycerol phosphate synthase (PGS) and phosphatidylserine synthase (PSS) levels in the absence or presence of the indicated concentrations of MgCl2 (n = 4). (J) Activity of recombinant PSS in the absence or presence of the indicated concentrations of MgCl2 (n = 3). (K, L) Synergy analysis for MgCl2 with balofloxacin (BLFX) for ΔplsB (K) and ΔpgpA (L) (n = 3). Synergy is represented using a color scale or an isobologram, which compares the dose needed for 50% inhibition for synergistic agents (blue) and non-synergistic (i.e., additive) agents (red). Results are displayed as means ± SD, and statistically significant differences are identified by Kruskal–Wallis followed by Dunn’s multiple comparison post hoc test unless otherwise indicated. *p<0.05 and **p<0.01.
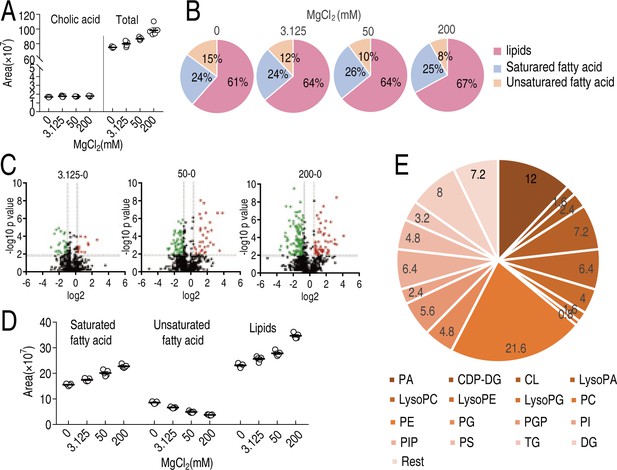
Lipidomes in the different concentrations of MgCl2.
(A) Area of fatty acids in the presence of indicated concentrations of MgCl2. (B) Percentage of lipids, saturated fatty acid, and unsaturated fatty acid in the presence of indicated concentration of MgCl2. (C) Volcano plots of lipidomics of indicated concentration of MgCl2 compared to non-treated control. (D) Relative abundance of saturated fatty acids, unsaturated fatty acids and lipids in the presence of indicated concentrations of MgCl2. (E) Relative percentage of indicated lipids.
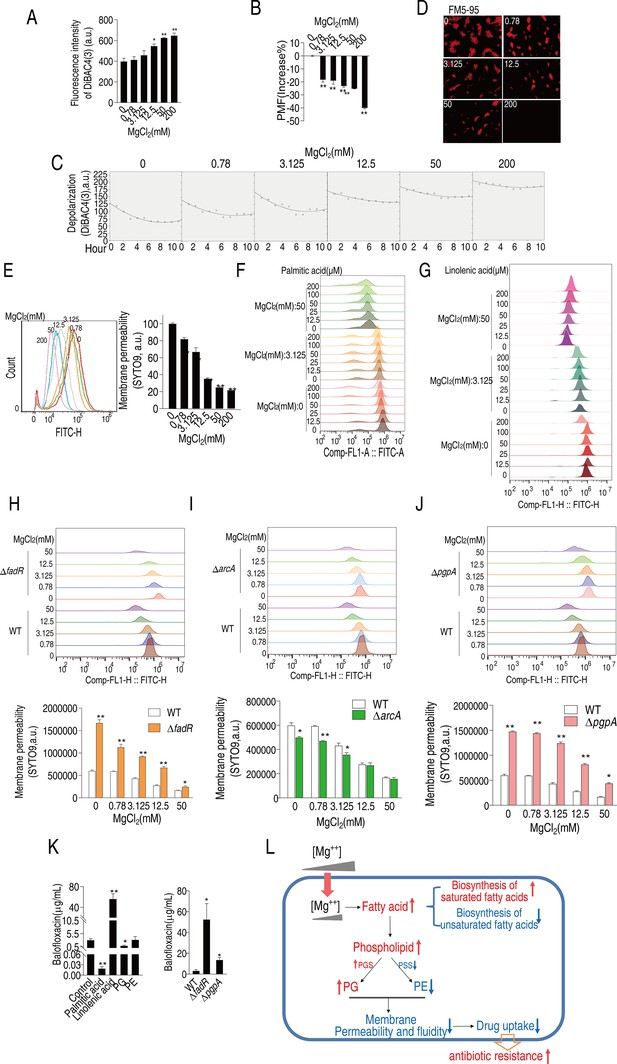
Mg2+ regulates membrane polarization, permeability, and fluidity to confer balofloxacin (BLFX) resistance.
(A, B) Depolarization (A) and proton motive force (PMF) (B) of ATCC33787 in the absence of or at the indicated concentrations of MgCl2 (n = 3). (C, D) Dynamic depolarization. (D) Membrane fluidity of ATCC33787 in the absence or presence of indicated concentrations of MgCl2, as shown by fluorescence microscopy. (E) Membrane permeability of ATCC33787 in the absence or presence of the indicated concentrations of MgCl2 (n = 3). (F, G) Membrane permeability of ATCC33787 cultured in palmitic acid (F) or linolenic acid (G) at the indicated concentrations of MgCl2 (n = 3). (H, I) Membrane permeability of ΔfadR (H) and ΔarcA (I) in the absence or presence of the indicated concentrations of MgCl2 (n = 3). (J) Membrane permeability of ΔpgpA in the absence or presence of the indicated concentrations of MgCl2 (n = 3). (K) Intracellular BLFX of ATCC33787 in the presence of palmitic acid, linolenic acid, phosphatidylglycerol (PG), and phosphatidylethanolamine (PE) (left panel) or ΔfadR and ΔpgpA mutants (right panel) (n = 3). (L) Diagram of mechanisms of Mg2+-mediated resistance to BLFX. Results are displayed as means ± SD, and statistically significant differences are identified by Kruskal–Wallis followed by Dunn’s multiple comparison post hoc test unless otherwise indicated. *p<0.05 and **p<0.01.
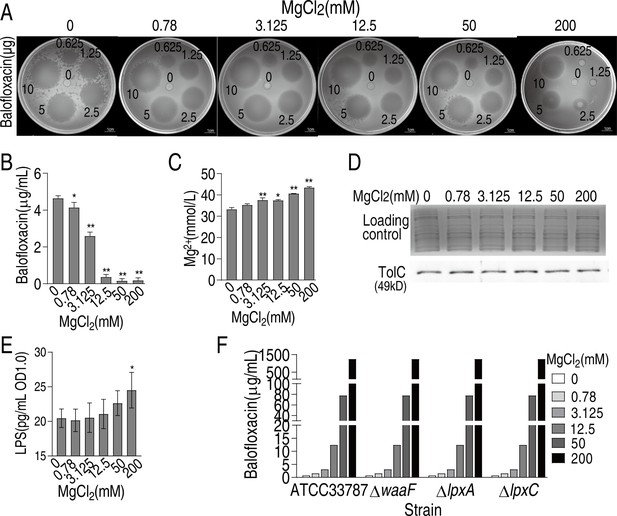
Mg2+ decreases balofloxacin (BLFX) uptake.
(A) Oxford cup test for effect of different concentrations of Mg2+ on antibacterial action of different dose of BLFX to ATCC33787. To do this, 0, 0.78, 3.125, 12.5, 50, and 200 mM MgCl2 were individually mixed with 0, 12.5, 25, 50, 100, or 200 μg/mL BLFX for 5 hr. Then, 50 μL were added into Oxford cup for antibacterial efficiency, which contained 0, 0.625, 1.25, 2.5, 5, or 10 μg BLFX, respectively. (B) Intracellular BLFX of ATCC 33787 in artificial seawater (ASWT) with the indicated concentrations of MgCl2 and 60 μg/mL BLFX. (C) Intracellular Mg2+ of ATCC 33787 in ASWT with the indicated concentrations of MgCl2. (D) Western blot for abundance of TolC in the presence of MgCl2. Whole-cell lysates resolved by SDS-PAGE gel were stained with Coomassie brilliant blue as loading control. (E) LPS quantification at the indicated concentrations of MgCl2. (F) MIC of ATCC 33787 and its mutants ΔwaaF、ΔlpxA、ΔlpxC in ASWT with the indicated concentrations of MgCl2, which is measured using the microtiter-dilution method.
-
Appendix 1—figure 1—source data 1
PDF file containing original western blots in 1 Appendix 1—figure 1, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100427/elife-100427-app1-fig1-data1-v1.zip
-
Appendix 1—figure 1—source data 2
Original files for western blot analysis displayed in Appendix 1—figure 1.
- https://cdn.elifesciences.org/articles/100427/elife-100427-app1-fig1-data2-v1.zip
Additional files
-
Source data 1
File containing original western blots in Appendix 1, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100427/elife-100427-data1-v1.zip
-
Source data 2
Original files for western blot analysis displayed in Appendix 1.
- https://cdn.elifesciences.org/articles/100427/elife-100427-data2-v1.zip
-
Supplementary file 1
Gradients of medium and primers used in this study.
(a) Comparison in components in LBS and ASWT. (b) Primes used in the present study. (c) Primers used in the present study for the construction of gene-deleted mutants.
- https://cdn.elifesciences.org/articles/100427/elife-100427-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/100427/elife-100427-mdarchecklist1-v1.pdf
-
Appendix 1—figure 1—source data 1
PDF file containing original western blots in 1 Appendix 1—figure 1, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100427/elife-100427-app1-fig1-data1-v1.zip
-
Appendix 1—figure 1—source data 2
Original files for western blot analysis displayed in Appendix 1—figure 1.
- https://cdn.elifesciences.org/articles/100427/elife-100427-app1-fig1-data2-v1.zip