A comparative study of the cryo-EM structures of Saccharomyces cerevisiae and human anaphase-promoting complex/cyclosome (APC/C)
Figures
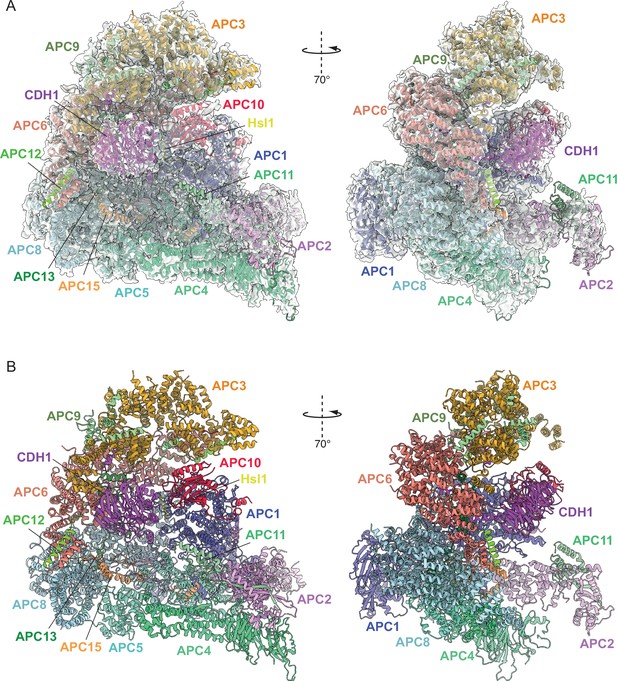
Overall structure of the APC/CCDH1:Hsl1 complex.
(A) Two views of the APC/CCDH1:Hsl1 ternary complex fitted into the 4.0 Å cryo-EM map. (B) Two views of the APC/CCDH1:Hsl1 ternary complex shown as ribbon representations.
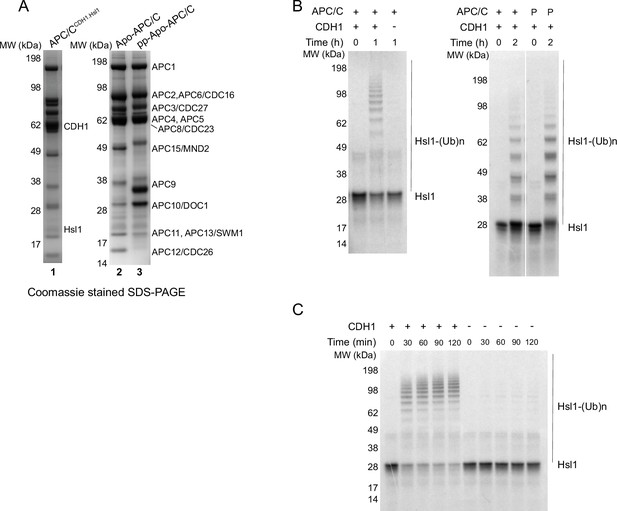
SDS PAGE gels of purified APC/C complexes and ubiquitylation assays.
(A) APC/CCDH1:Hsl1 complex, unphosphorylated apo-APC, phosphorylated (pp) apo-APC (lanes 1–3). (B) Ubiquitylation assay showing activation by CDH1. Phosphorylation (P: phosphorylated APC/C) did not significantly affect activity. (C) Time course of ubiquitylation reaction.
-
Figure 1—figure supplement 1—source data 1
PDF file of original Coomassie stained SDS PAGE gel for Figure 1—figure supplement 1A and Western blots for Figure 1—figure supplement 1B and C, with no labels.
- https://cdn.elifesciences.org/articles/100821/elife-100821-fig1-figsupp1-data1-v1.zip
-
Figure 1—figure supplement 1—source data 2
PDF file of original Coomassie stained SDS PAGE gel for Figure 1—figure supplement 1A and Western blots for Figure 1—figure supplement 1B and C, indicating relevant lanes (boxed in red) and treatments.
- https://cdn.elifesciences.org/articles/100821/elife-100821-fig1-figsupp1-data2-v1.zip
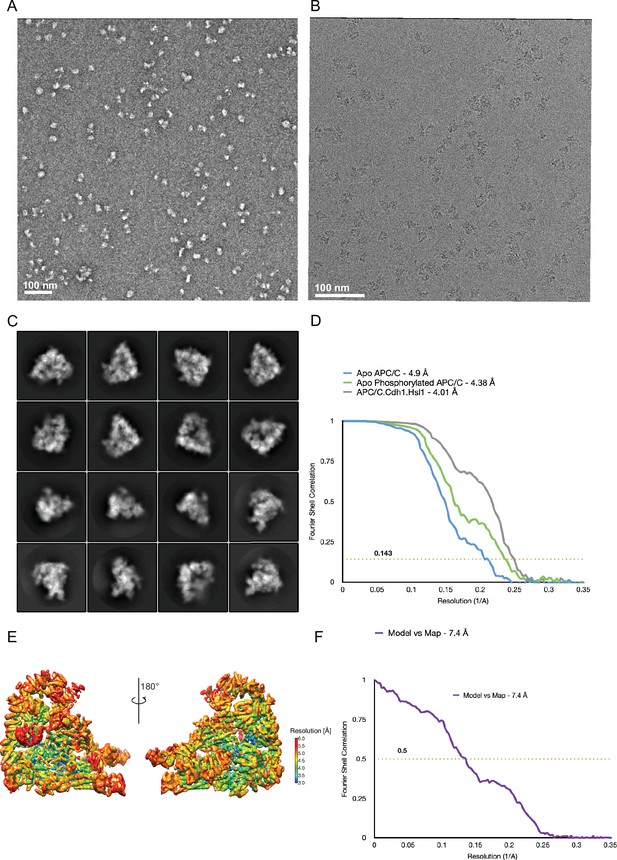
EM images and 2D class averages of APC/CCDH1:Hsl1 complex.
(A) Representative negative stain image of the APC/CCDH1:Hsl1 complex. (B) Representative cryo-EM image of the APC/CCDH1:Hsl1 complex. (C) Gallery of 2D-class averages of APC/CCDH1:Hsl1. (D) Fourier Shell Correlation (FSC) plots of the main maps used to generate molecular models APC/CCDH1:Hsl1, unphosphorylated apo-APC/C and phosphorylated apo-APC/C. (E) Local resolution map of APC/CCDH1:Hsl1. (F) Fourier Shell Correlation (FSC) plots of the main maps used to generate molecular models.
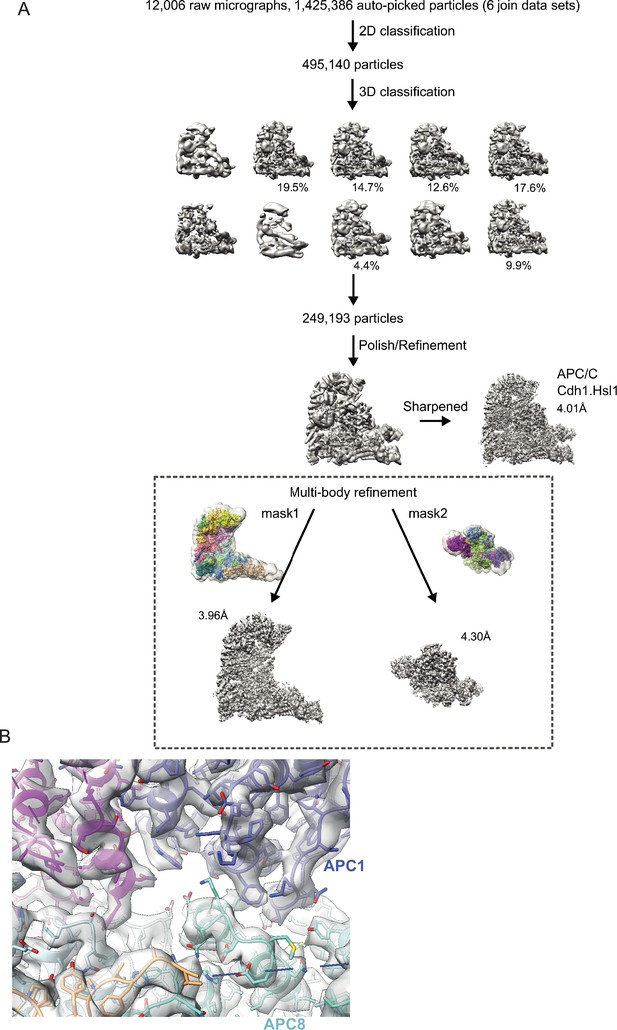
Data processing pipeline for APC/CCDH1:Hsl1 complex cryo-EM reconstructions.
(A) Cryo-EM data processing workflow summary for the APC/CCDH1:Hsl1 complex as described in the Methods section. (B) Example of cryo-EM map with fitted coordinates.
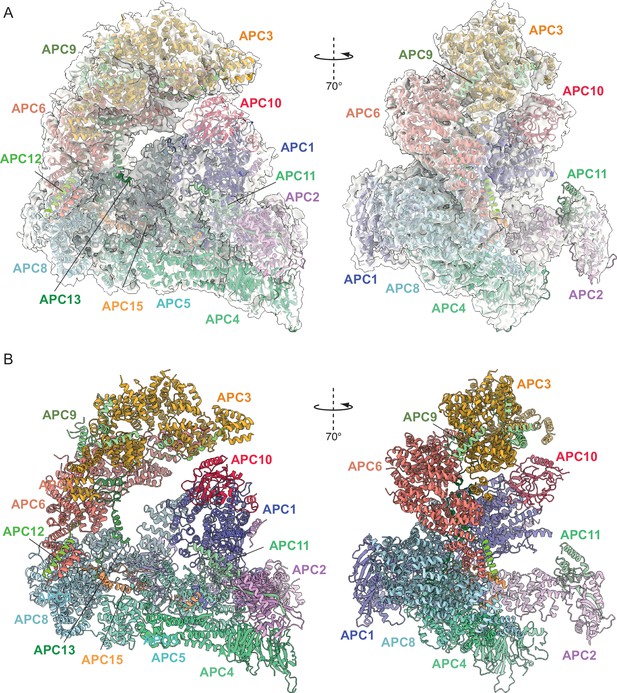
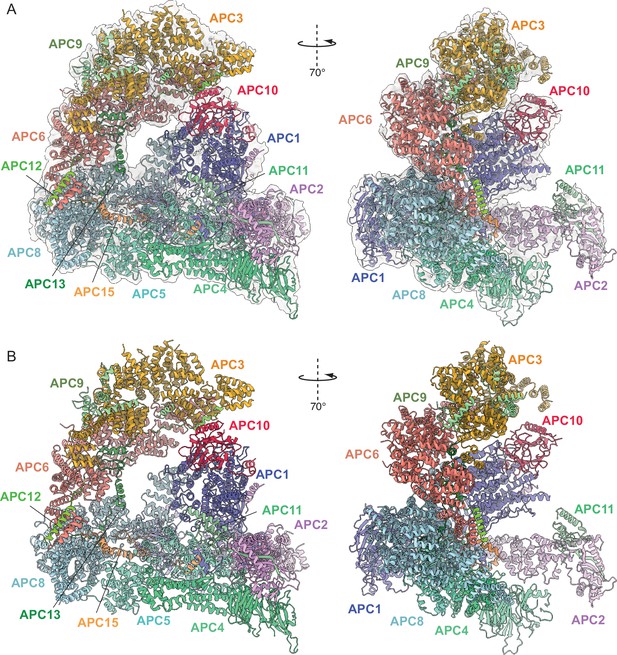
Overall structure of unphosphorylated apo-APC/C.
(A) Two views of apo-APC/C fitted into the 4.9 Å cryo-EM map. (B) Two views of apo-APC/C shown as ribbon representations.
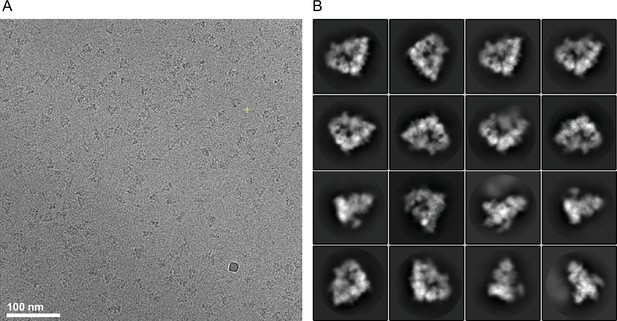
Cryo-EM images and 2D class averages of apo-APC/C complexes.
(A) Representative cryo-EM image of unphosphorylated apo-APC/C. (B) Gallery of 2D-class averages of unphosphorylated apo-APC/C.
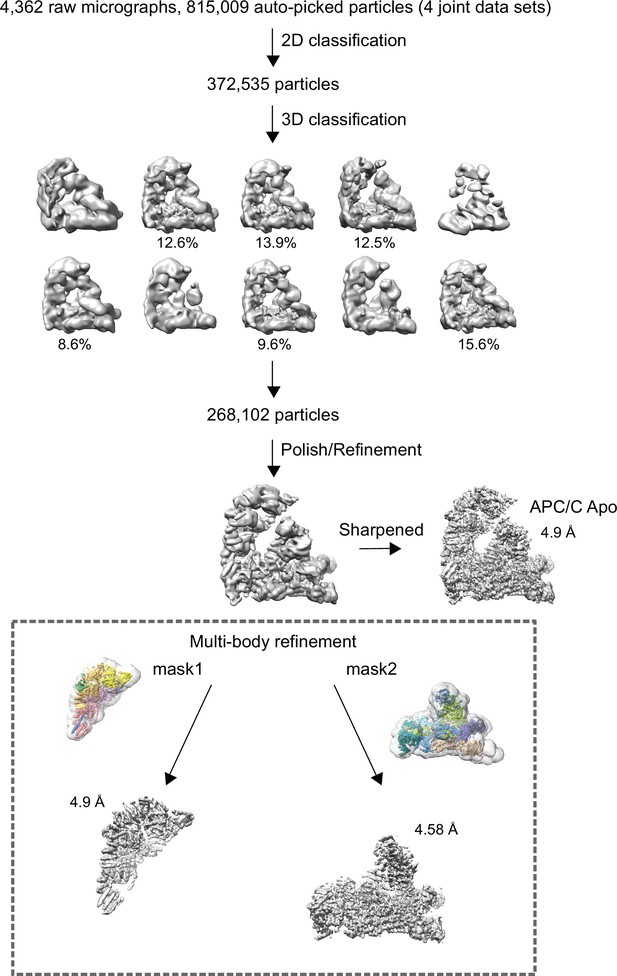
Data processing pipeline for unphosphorylated apo-APC/C cryo-EM reconstructions.
Cryo-EM data processing workflow summary for unphosphorylated apo-APC/C as described in the Materials and methods section.
Overall structure of phosphorylated apo-APC/C.
(A) Two views of phosphorylated apo-APC/C fitted into the 4.5 Å cryo-EM map. (B) Two views of apo-APC/C shown as ribbon representations.
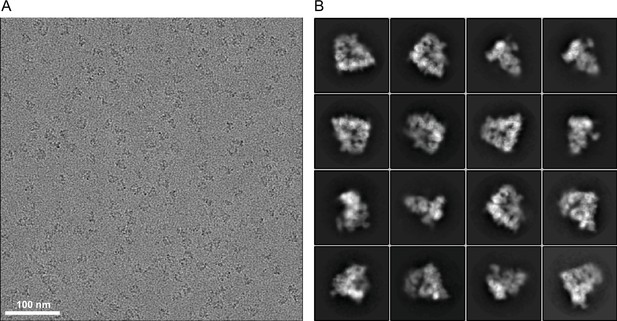
Cryo-EM images and 2D class averages of phosphorylated apo-APC/C complexes.
(A) Representative cryo-EM image of phosphorylated apo-APC/C. (B), Gallery of 2D-class averages of phosphorylated apo-APC/C.
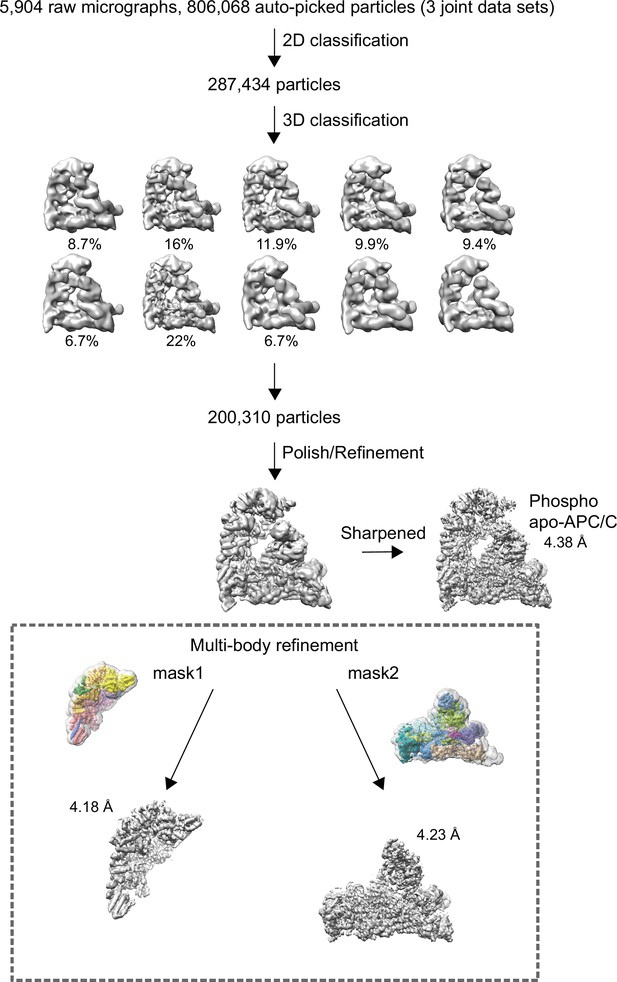
Data processing pipeline for phosphorylated apo-APC/C cryo-EM reconstructions.
Cryo-EM data processing workflow summary for phosphorylated apo-APC/C as described in the Materials and methods section.
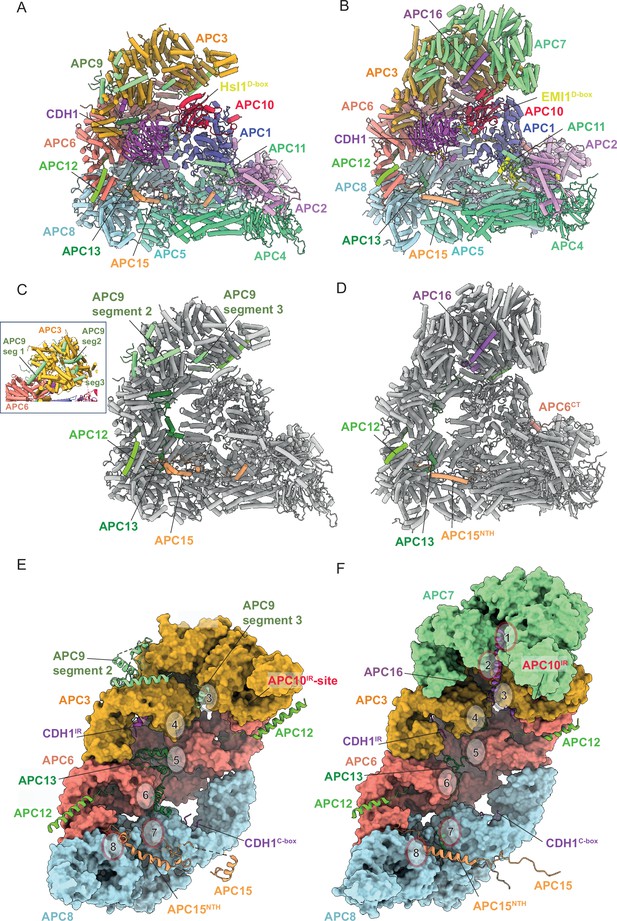
Comparison of S. cerevisiae APC/CCDH1:Hsl1 with human APC/CCDH1:EMI1.
(A) S. cerevisiae and (B) human complexes subunits colour-coded. (C) S. cerevisiae and (D) human complexes with large subunits coloured in grey and small IDP subunits (APC9, APC12, APC13, APC15, APC16) colour-coded. The structure of human APC/CCDH1:EMI1 from Höfler et al., 2024 (PDB 7QE7). (E) and (F) four small IDP-subunits (APC9, APC13, APC15, APC16) contribute to interacting with equivalent, quasi-symmetrical sites on the outer surfaces of the TPR subunits of the TPR lobe. TPR lobes of S. cerevisiae (E) and human APC/C (F) are depicted as a surface representations. The contact sites with the three small subunits that contact the S. cerevisiae TPR lobe are numbered 3–8 after human APC/C (Chang et al., 2015) that has sites 8 sites due to APC7.
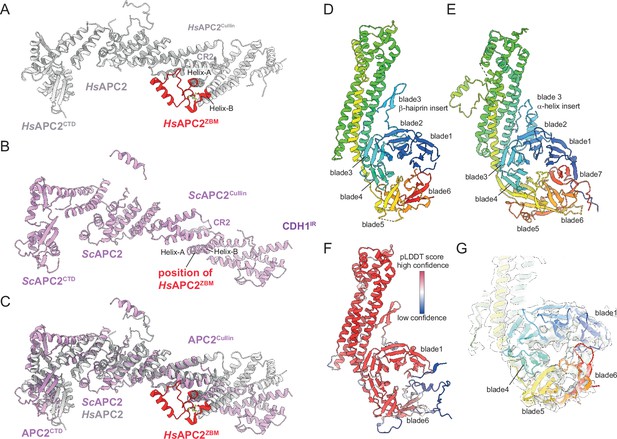
Comparison of S. cerevisiae and human APC2 and APC4 subunits.
(A) Human APC2 with APC2ZBM of CRL2 coloured red. (B) S. cerevisiae APC2. The position of human APC2ZBM is indicated. (C) Superimposition S. cerevisiae APC2 (purple) with human APC2 (grey) on CRL2. Human APC2 from Höfler et al., 2024 (PDB 7QE7). (D) S. cerevisiae APC4, colour-coded from N- to C-termini with a blue to red ramp. (E) Human APC4, colour-coded from N- to C-termini with a blue to red ramp. S. cerevisiae APC4WD40 is a six-bladed β-propeller in contrast to the seven bladed β-propeller of human APC4WD40. (F) AlphaFold2 prediction of S. cerevisiae APC4 colour-coded according to pLDDT score. No seventh bade is predicted. The α-helix in place of the seventh blade is predicted with low confidence. (G) Fit of S. cerevisiae APC4WD40 into the APC/CCDH1:Hsl1 cryo-EM map showing lack of density for a seventh blade. Human APC4 from Höfler et al., 2024 (PDB 7QE7).
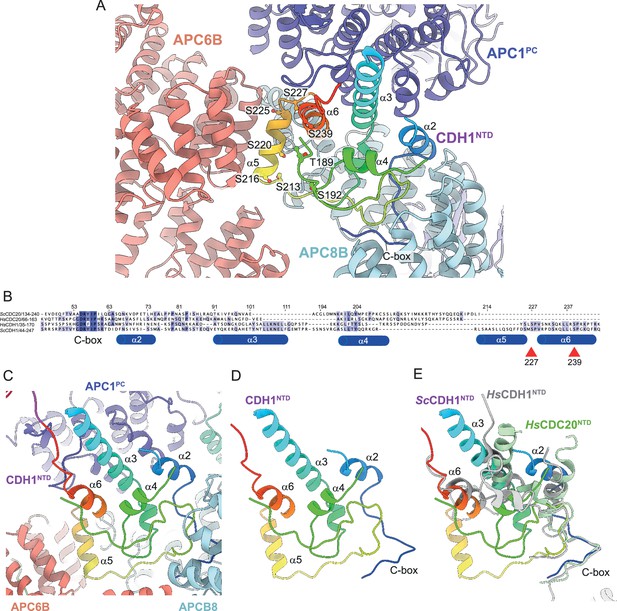
CDH1NTD contacts to APC/C and control by phosphorylation.
(A) In S. cerevisiae CDH1, α2, α3 and α6 contact the PC domain of APC1 (APC1PC) (conserved with human CDH1). α2 also contacts APC8B. α5, unique to S. cerevisiae CDH1, forms extensive contacts to APC6B. Human CDC20NTD (light green) shares α2 and α3 with CDH1NTD. (B) MSA of S. cerevisiae and human CDH1NTD and CDC20NTD showing α-helices α2 to α6 of S. cerevisiae CDH1NTD and the conserved sites of CDK phosphorylation on CDH1 (red arrows). (C) S. cerevisiae CDH1NTD colour-coded from N- to C-termini with a blue to red ramp interacting with APC1PC, APC6B and APC8B. (D) Same view as (c) without APC1PC, APC6B and APC8B. (E) Superimposition of S. cerevisiae CDH1NTD coloured blue-to-red, human CDH1NTD (grey) (from Höfler et al., 2024, PDB 7QE7) and human CDC20NTD (light green) (from Zhang et al., 2019 PDB 6Q6G). α-helices α2, α3 and α6 are conserved in all three structures.
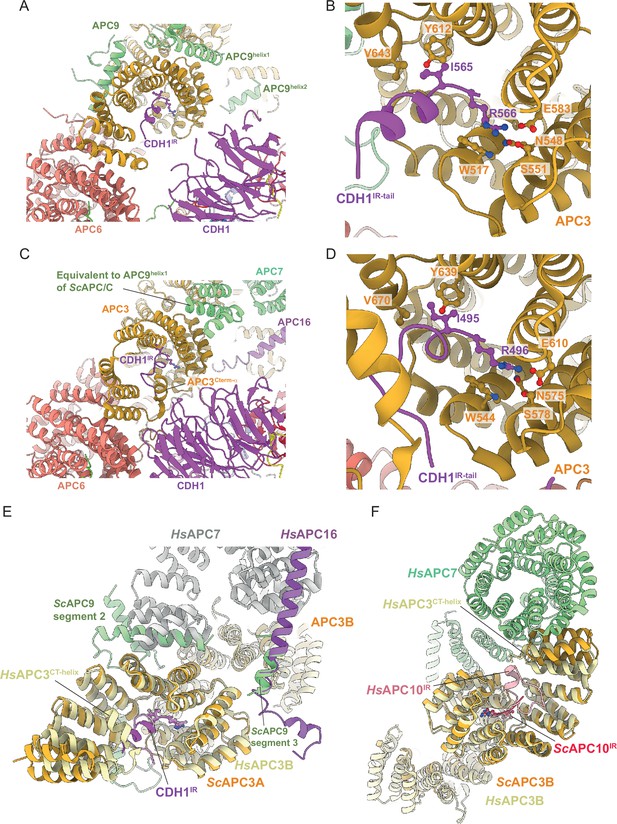
Comparison of CDH1IR binding to S.cerevisiae and human APC/CCDH1 complexes.
(A) Overall view of the CDH1IR-binding site on S. cerevisiae APC/CCDH1:Hsl1. (B) Zoomed view of this complex. (C) Overall view of the CDH1IR-binding site on human APC/CCDH1:EMI1. (D) Zoomed view of this complex. The structure of human APC/CCDH1:EMI1 from Höfler et al., 2024 (PDB 7QE7). (E, F) APC9 of S. cerevisiae APC/C partially mimics APC7 and APC16 of human APC/C. (E) In S. cerevisiae APC/C, the interaction of APC9 with APC3A (segment 2 of APC9) and APC3B (segment 3 of APC9) partially mimics the respective APC7 and APC16 interactions with human APC3. APC9 segment 2 interactions with the C-terminal TPR helix of APC3A may stabilise the CDH1IR-binding site of APC3A. (F) Similarly, in human APC/C, the interface of APC7B with APC3B might function to stabilise the APC10IR-binding site on APC3B. Human APC/CCDH1:EMI1 from Höfler et al., 2024 (PDB 7QE7).
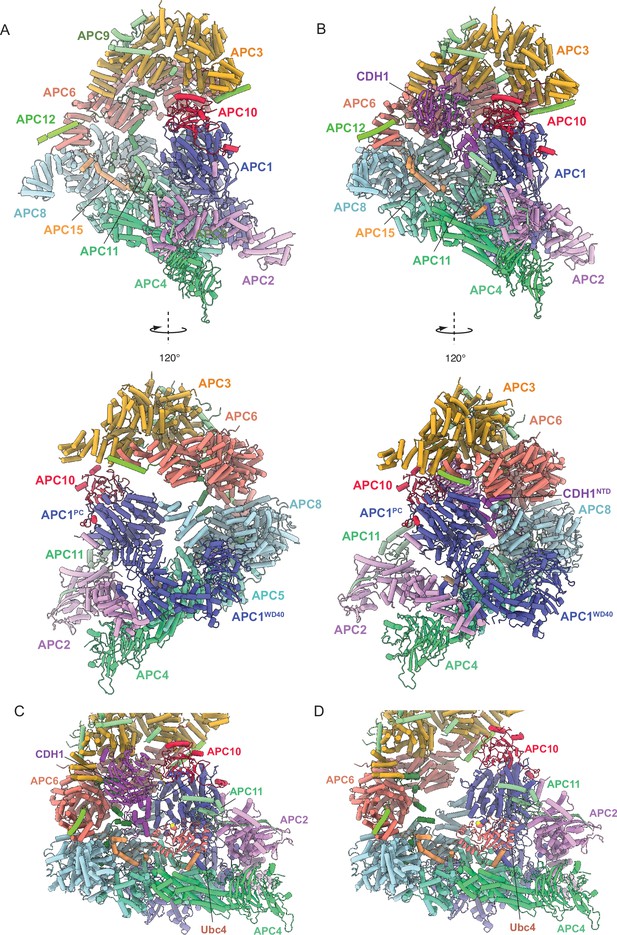
Comparing apo-APC/C and the APC/CCDH1:Hsl1 complex shows both apo-APC/C and the APC/CCDH1:Hsl1 complexes are competent to bind the Ubc4.
(A) Two views of apo-APC/C. (B) Two views of APC/CCDH1:Hsl1. The coordinates were superimposed on APC1PC. In the apo-APC/C complex, there are no contacts between APC1PC of the platform module and APC6B and APC8B of the TPR lobe. In the APC/CCDH1:Hsl1, CDH1NTD bridges APC1PC with APC6B and APC8B. The overall conformations are similar, specifically the APC2:APC11 catalytic module adopts a raised conformation in both states. (C) Model of APC/CCDH1:Hsl1 in complex with Ubc4. (D) Model of apo-APC/C in complex with Ubc4. The position of the APC2:APC11 catalytic module in apo-APC/C does not exclude Ubc4 binding. Model of APC11:E2 based on the S. cerevisiae Not4 RING:Ubc4 complex (PDB 5AIE; Bhaskar et al., 2015).
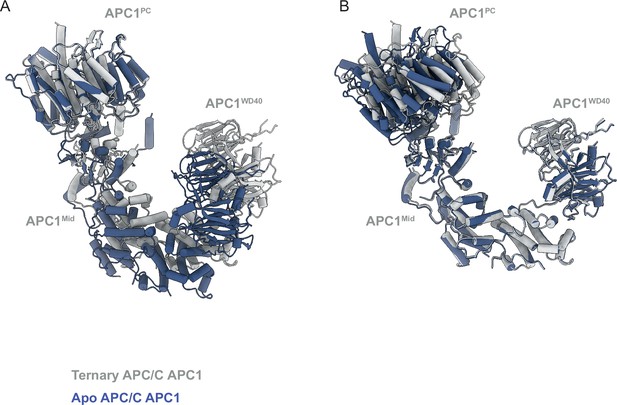
Conformational change of APC1 on conversion from apo-APC/C to APC/CCDH1:Hsl1.
(A) Superimposed on APC1PC. (B) Superimposed on the combined APC1Mid-APC1WD40 domains. On conversion, there is a small tilt of APC1PC relative to the combined APC1Mid-APC1WD40 domains that form a rigid unit.
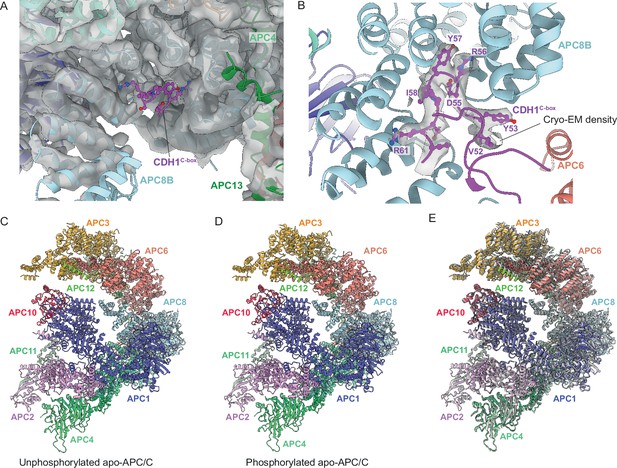
Regulation of APC/C by phosphorylation.
The C-box binding site is un-obstructed in unphosphorylated apo-APC/C. (A) Cryo-EM density from the unphosphorylated apo-APC/C map corresponding to the C-box binding site of APC8B with the fitted CDH1C-box modelled from APC/CCDH1:Hsl1. We observe no density at the APC8B C-box binding site. (B) Cryo-EM map density for the CDH1C-box from the APC/CCDH1:Hsl1 reconstruction with fitted coordinates. (C–E) Comparison of unphosphorylated apo-APC/C and phosphorylated apo-APC/C: (C) Unphosphorylated apo-APC/C. (D) Phosphorylated apo-APC/C. (E) Superimposition of unphosphorylated apo-APC/C (coloured by subunit assignment) and phosphorylated apo-APC/C (grey). There is a small relative shift of the TPR lobe. The coordinates were superimposed on APC1PC.
Videos
CDH1-induces conformational changes in APC/C.
The video shows a morph of apo-APC/C to the ternary APC/CCDH1:Hsl1 complex indicating the conformational changes induced in APC/C as a consequence of CDH1 binding. The two conformational states were superimposed on the N-terminus of APC1. On transition to the ternary APC/CCDH1:Hsl1 state, APC3 and APC6 move closer to the APC1PC domain, as a result of the CDH1 N-terminal domain binding at the interface of APC1PC and APC6. The catalytic module of APC2-APC11 does not change conformation.
Tables
Cryo-EM data collection, refinement, and validation statistics.
| Apo-APC/C(EMD-15199)(PDB 8A5Y) | Phosphorylated apo-APC/C (EMD-15201)(PDB 8A61) | APC/CCDH1:Hsl1(EMD-15123)(PDB 8A3T) | |
|---|---|---|---|
| Data collection | |||
| EM | FEI Titan Krios | FEI Titan Krios | FEI Titan Krios |
| Detector | FEI Falcon Ill | FEI Falcon Ill | FEI Falcon Ill |
| Magnification | 59,000 | 59,000 | 59,000 |
| Voltage (keV) | 300 | 300 | 300 |
| Electron exposure (e-/Å2) | 59 | 59 | 59 |
| Defocus range (µm) | 2.6–3.9 | 2.6–3.9 | 2.6–3.9 |
| Pixel size (Å) | 1.38 | 1.38 | 1.38 |
| Reconstruction | |||
| Software | RELION 3.1 | RELION 3.1 | RELION 3.1 |
| Symmetry imposed | C1 | C1 | C1 |
| Initial particle images (N) | 815,009 | 806,068 | 1,425,386 |
| Final particle images (N) | 268,102 | 200,310 | 249,193 |
| Accuracy of rotations (°) | 1.76 | 1.79 | 1.16 |
| Accuracy of translations (°) | 1.01 | 0.93 | 0.62 |
| Map resolution (Å) | 4.9 | 4.4 | 4.0 |
| FSC threshold | 0.143 | 0.143 | 0.143 |
| Refinement | |||
| Software | Phenix | ||
| Resolution limit (Å) | 4.9 | 4.4 | 4.0 |
| RMSD bond length (Å) | 0.004 | 0.003 | 0.004 |
| RMSD bond angle (°) | 0.878 | 0.641 | 0.754 |
| Model to map fit | |||
| CC_mask | 0.756 | 0.793 | 0.752 |
| CC_volume | 0.748 | 0.785 | 0.750 |
| Validation | |||
| All-atom clash score | 18.46 | 15.14 | 18.84 |
| Ramachandran plot | |||
| Preferred (%) | 95.76 | 95.74 | 95.93 |
| Allowed (%) | 3.94 | 4.02 | 3.90 |
| Outliers (%) | 0.34 | 0.24 | 0.17 |
Additional files
-
Supplementary file 1
Tables of APC/C phosphorylation sites and ordered and disordered regions of APC/C subunits.
(a) Table of CDK Phosphorylation sites of S. cerevisiae APC/C. (b) Ordered and disordered regions of S. cerevisiae APC/C subunits
- https://cdn.elifesciences.org/articles/100821/elife-100821-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/100821/elife-100821-mdarchecklist1-v1.pdf





