Coordinated regulation of chemotaxis and resistance to copper by CsoR in Pseudomonas putida
Figures
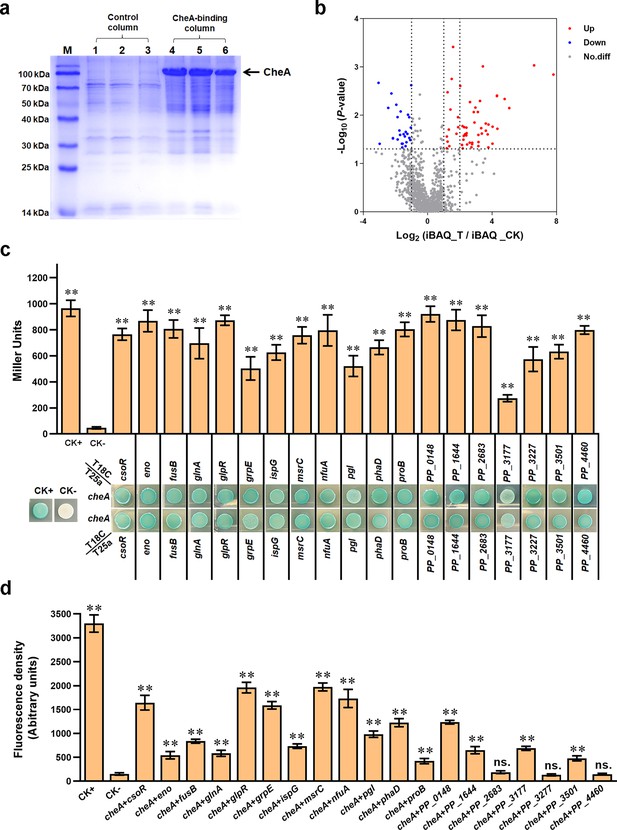
Screening and verifying proteins interacting with CheA.
(a) Protein samples obtained in pull-down assay and detected by SDS/PAGE. The ‘bait’ protein CheA on the gel was indicated. Lanes 1, 2, and 3 are samples from the control column, and lanes 4, 5, and 6 are samples from the CheA-binding column. M represents a protein marker. (b) The volcano plot shows the p-value and fold-change of all proteins identified in MS analyses. Red spots represent proteins that showed two or higher folds in the experimental group compared with the control group (p<0.05). Blue spots represent proteins with a higher amount in the control group (p<0.05). Grey spot proteins showed no apparent difference between the two groups (p>0.05). (c) Detect the interaction between CheA and indicated proteins via BTH. Blue indicates protein-protein interaction in the colony after 60 hr of incubation, while white indicates no protein-protein interaction. A colony containing T25a-zip and T18C-zip plasmids was used as a positive control (CK+), and a colony containing empty T25a and T18C plasmids was used as a negative control (CK-). The LacZ activities of colonies were shown above the colonies. (d) The red fluorescence intensities in BiFC assay. The results in panels c and d are the average of three independent assays. Error bars represent standard deviations. The asterisks above the column denote significant differences (**p<0.01) between indicated strains and CK- strain. ‘ns.’ represents none statistically significant between indicated strain and CK- strain.
-
Figure 1—source data 1
Excel file containing original SDS-PAGE gel for Figure 1a.
- https://cdn.elifesciences.org/articles/100914/elife-100914-fig1-data1-v1.zip
-
Figure 1—source data 2
Excel file containing original SDS-PAGE gel for Figure 1a, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100914/elife-100914-fig1-data2-v1.zip
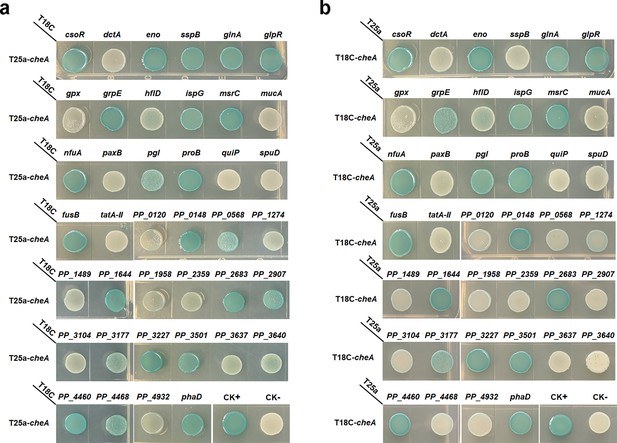
Detect the interaction between CheA and the 40 proteins using BTH.
CheA was cloned into T25a (a) and T18C (b). Blue indicates protein-protein interaction, and white indicates no interaction in the colony after 60 hr of incubation. A colony containing T25a-zip and T18C-zip plasmids was used as a positive control (CK+), and a colony containing empty T25a and T18C plasmids was used as a negative control (CK-).
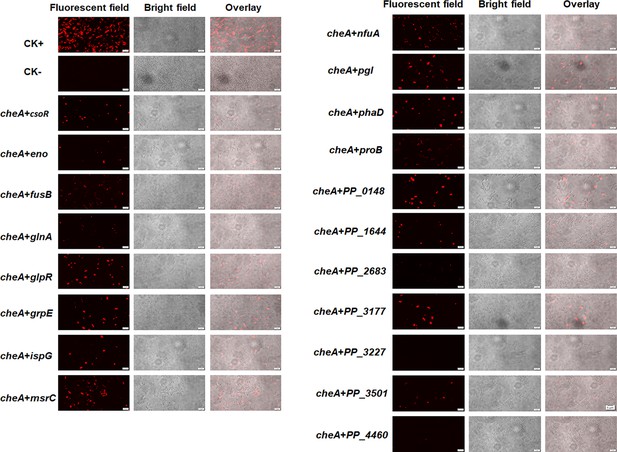
Detect the interaction between CheA and indicated proteins using BiFC.
Representative images for each pair of proteins were shown. Jun-KN151 +Fos-LC151 and KN151 +LC151 were used as CK +and CK-, respectively. RFP channel images (fluorescent field), bright field images, and overlay images of the same field are shown. Scale bar = 2 μm.
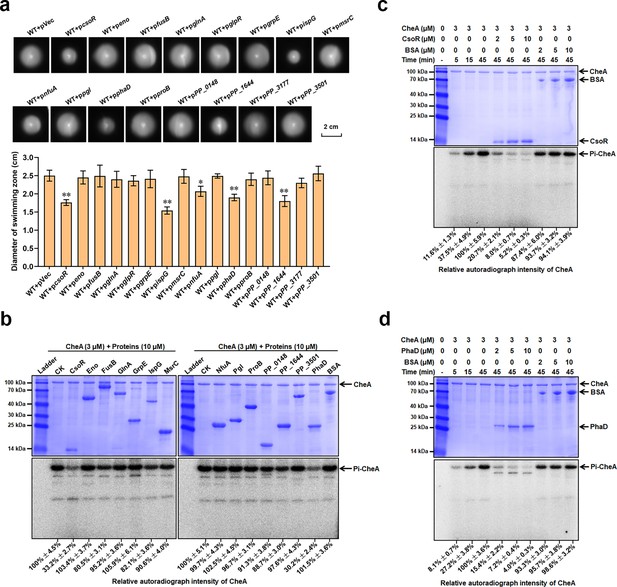
CsoR and PhaD inhibit CheA autophosphorylation.
(a) Chemotaxis of indicated strains on semisolid plates. Photos of colonies on the top were taken after 16 hr incubation at 28 °C. Diameters of colonies (swimming zone) shown at down were calculated from three replicates. The asterisks above the column denote significant differences (*p<0.05, **p<0.01) between indicated strains and control strain (WT + pVec) analyzed by Student’s t-test. (b) Effect of the 14 proteins on the CheA autophosphorylation. The name/ID and the concentration of tested proteins added in each lane are indicated above the gel. CK represents CheA alone in the reaction mixture. BSA is used as a negative control. The ladder represents a protein marker. (c and d) CsoR (c) and PhaD (d) affect CheA autophosphorylation. The concentration of tested proteins added in each lane is indicated above the gel. The time represents the time of the CheA autophosphorylation reaction. The SDS-PAGE gels in panels b, c, and d were detected by Coomassie Blue Staining (Above) and autoradiograph (Below). The experiments for panels b, c, and d were repeated three times, and a representative photo was shown. The relative autoradiograph intensity of the CheA band was calculated using Image J software and shown below each lane.
-
Figure 2—source data 1
Excel file containing original SDS-PAGE gels and autoradiograph photos for Figure 2b, c and d.
- https://cdn.elifesciences.org/articles/100914/elife-100914-fig2-data1-v1.zip
-
Figure 2—source data 2
Excel file containing original SDS-PAGE gels and autoradiograph photos for Figure 2b, c and d, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100914/elife-100914-fig2-data2-v1.zip
Growth curve of the 16 overexpression strains and wild-type strain in liquid LB broth (100 mL in a 250 mL triangular glass flask, at 28 °C with 180 rpm shaking).
The optical density at 600 nm (OD600) was used to characterize the growth of bacterial cells in the medium.
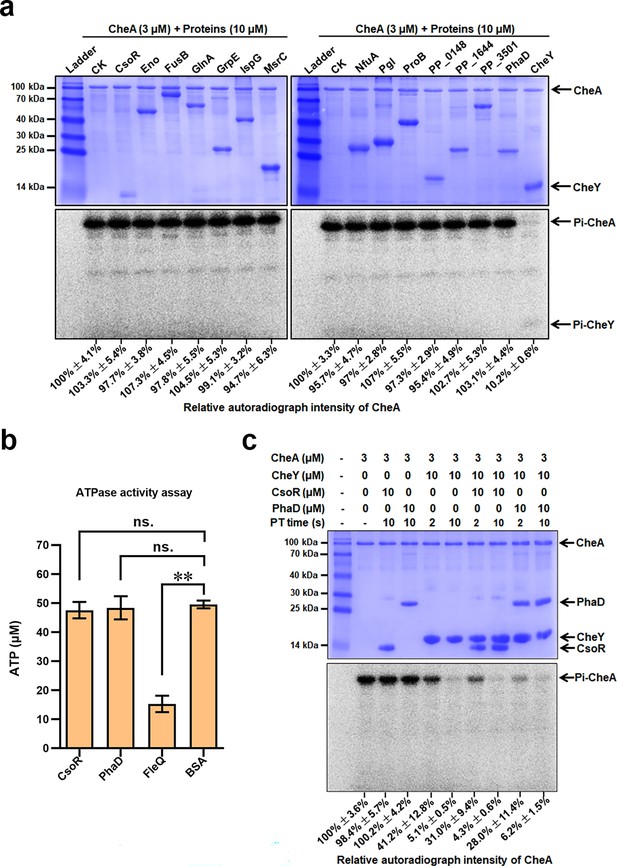
Role of target proteins in the CheA-mediated transphosphorylation.
(a) Transphosphorylation between CheA and the 14 proteins. Target proteins were added to the phosphorylated CheA and incubated for 10 s before adding termination buffer. CheY was added as a positive control. (b) ATPase activity of indicated proteins. The amount of remaining ATP after incubation with 10 μM indicated proteins for 30 min. BSA was used as a negative control. The asterisks represent statistically significant differences between FleQ and BSA (**p<0.01). ‘ns.’ represents no statistically significant between the indicated protein and BSA. (c) Effect of CsoR and PhaD on the transphosphorylation between CheA and CheY. The PT time in seconds (2 s and 10 s) represents the time of transphosphorylation. The bands of CheA, CheY, CsoR, and PhaD on the gel were indicated with arrows. The relative autoradiograph intensity of the CheA band was calculated using Image J software, shown below each lane.
-
Figure 2—figure supplement 2—source data 1
Excel file containing original SDS-PAGE gels and autoradiograph photos for Figure 2—figure supplement 2a and c.
- https://cdn.elifesciences.org/articles/100914/elife-100914-fig2-figsupp2-data1-v1.zip
-
Figure 2—figure supplement 2—source data 2
Excel file containing original SDS-PAGE gels and autoradiograph photos for Figure 2—figure supplement 2a and c, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100914/elife-100914-fig2-figsupp2-data2-v1.zip
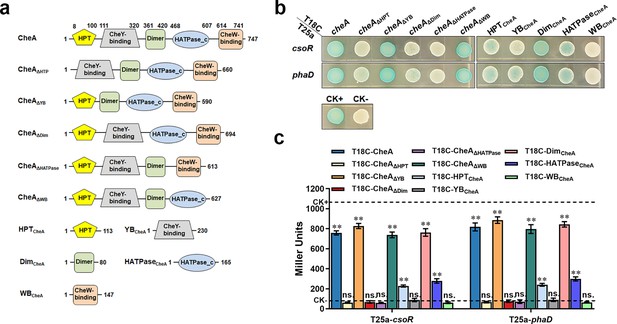
CheA domains involved in interacting with CsoR and PhaD.
(a) Schematic diagram of CheA and truncated CheA proteins. The predicted domains are based on the Pfam database and the amino acid positions where the predicted domains start and end are shown. (b) The interaction between CheA domains and CsoR/PhaD was tested using BTH. Blue indicates protein-protein interaction in the colony after 60 h of incubation, while white indicates no protein-protein interaction. A colony containing T25a-zip and T18C-zip plasmids was used as a positive control (CK+), and a colony containing empty T25a and T18C plasmids was used as a negative control (CK-). (c) Confirmation of BTH interactions in panel B by LacZ activity assay. The results are the average of three independent assays. Error bars represent standard deviations. The asterisks above the column denote significant differences (**p<0.01) between indicated strains and CK- strain analyzed by Student’s t-test. ‘ns.’ represents none statistically significant between indicated strain and CK- strain.
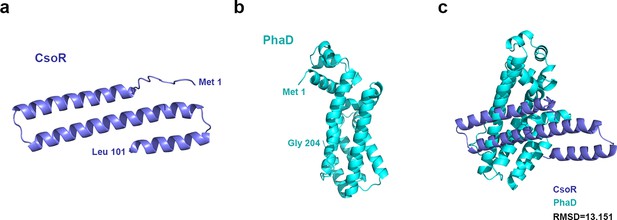
Predicted structure of the P. putida CsoR.
(a) and PhaD (b) obtained using an online AlphaFold server (https://www.alphafold.ebi.ac.uk). Position of the first and the last amino acid residue of CsoR and PhaD are indicated on the structure. (c) The structure alignment of CsoR and PhaD was performed with PyMOL software. The protein name and its structure were shown with the same color. The differences between protein structures were quantified using the Root Mean Square Deviation (RMSD) index. An RMSD value lower than 2 was considered a high similarity between two compared proteins.
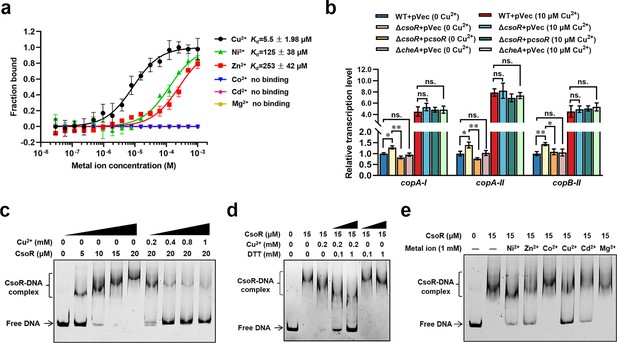
CsoR is a metal-binding repressor.
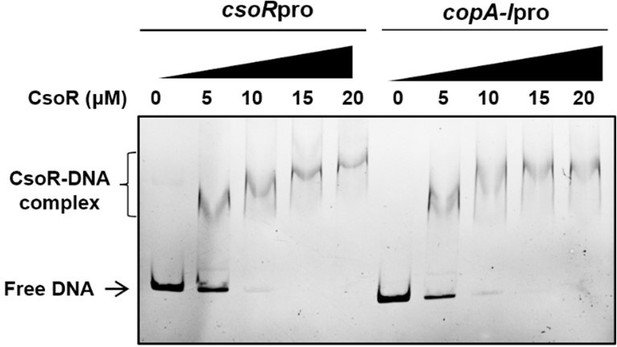
(a) MST analysis of the interaction between CsoR-GFP and metal ions. CsoR-GFP (250 nM) was incubated with increasing concentrations of metal ions. (b) Analysis of relative transcription level of target genes in wild-type (WT +pVec), csoR mutant (ΔcsoR +pVec), complemented strain (ΔcsoR +pcsoR), and cheA mutant (ΔcheA +pVec) in the presence and absence of CuCl2 (10 μM) by qRT-PCR. The results are the average of three independent assays. Error bars represent standard deviations. The asterisks represent statistically significant differences between the two compared strains (*p<0.05, **p<0.01). ‘ns.’ represents none statistically significant between two compared strains. (c) Analysis for interactions between CsoR and copA-I promoter DNA using EMSA. (d) The effect of DTT/CuCl2 +DTT on the interaction between CsoR and copA-I promoter DNA. (e) The effect of indicated metal ions on the interaction between CsoR and copA-I promoter DNA. The concentrations of CsoR, metal ions, and DTT in panels c, d, and e added in each lane are shown above the gel. Free DNA and CsoR-DNA complex are indicated.
-
Figure 4—source data 1
Excel file containing original EMSA Native-PAGE gels for Figure 4c, d and e.
- https://cdn.elifesciences.org/articles/100914/elife-100914-fig4-data1-v1.zip
-
Figure 4—source data 2
Excel file containing original EMSA Native-PAGE gels for Figure 4c, d and e, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100914/elife-100914-fig4-data2-v1.zip
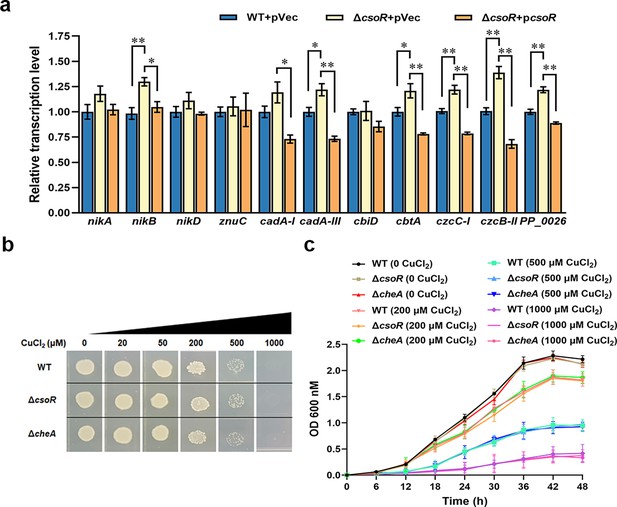
Function of CsoR in the expression of metal resistant genes and the bacterial growth under copper stress.
(a) Analysis of relative transcription level of metal resistant genes in wild-type (WT + pVec), csoR mutant (ΔcsoR +pVec), and complemented strain (ΔcsoR + pcsoR) by qRT-PCR. The results are the average of three independent assays. Error bars represent standard deviations. The asterisks represent statistically significant differences between the two compared strains (*p<0.05, **p<0.01). (b and c) Effect of csoR/cheA deletion on bacterial growth under copper stress. Growth of WT, ΔcsoR, and ΔcheA on M9 minimal medium agar plate (b) and in liquid 1/4 LB medium (c) containing different CuCl2 concentrations. A same amount of bacterial culture (OD600=0.5) was spotted onto the plate and incubated at 28 °C for 24 hr. The concentrations of CuCl2 used in the assay were indicated above.
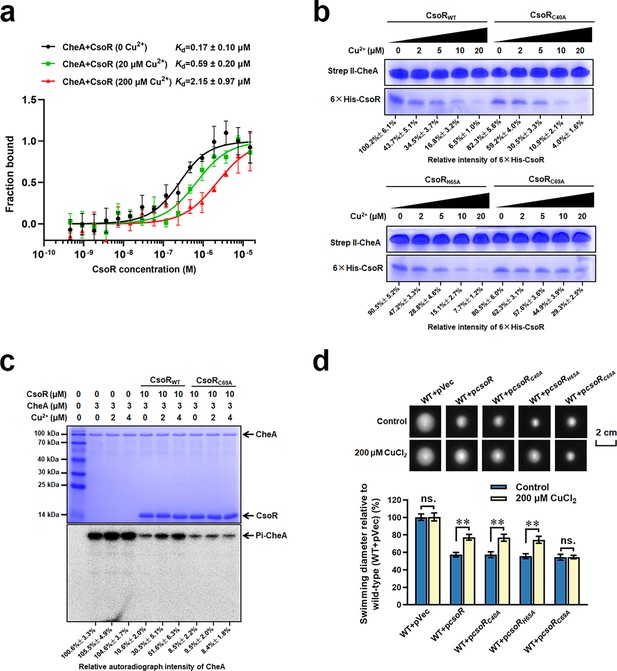
Copper inhibits the interaction between CheA and CsoR.
(a) MST analysis of the interaction between CheA-GFP and CsoR in the presence of Cu2+. CheA-GFP (160 nM) was incubated with increasing concentrations of CsoR. (b) SDS-PAGE detected protein samples obtained in a pull-down assay. The ‘bait’ protein Strep-CheA and the ‘prey’ protein His-CsoR on the gel were indicated. The gel showed the influence of Cu2+ on the amount of ‘prey’ protein His-CsoR. The concentration of Cu2+ added in each pull-down assay was displayed above the gel. The SDS-PAGE gel was detected by Coomassie Blue Staining. (c) CheA autophosphorylation in the presence of CsoR and Cu2+. The tested proteins and Cu2+ concentrations added in each lane are indicated above the gel. The SDS-PAGE gel was detected by Coomassie Blue Staining (Above) and autoradiograph (Below). The relative intensity of the CsoR band in panel b and the autoradiograph intensity of the CheA band in panel c were calculated using Image J software and shown below each lane. (d) Chemotaxis of indicated strains on semisolid plates supplied with or without CuCl2. Photos of colonies on the top were taken after 16 hr (for the control plate) or 18 hr (for the copper-adding plate) incubation at 28 °C. The diameters of colonies were measured and normalized to the diameters of WT + pVec, shown below. The asterisks above the column denote significant differences (**p<0.01) between two indicated strains analyzed by Student’s t-test. ‘ns.’ represents none statistically significant between two compared strains.
-
Figure 5—source data 1
Excel file containing original SDS-PAGE gels and autoradiograph photo for Figure 5b and c.
- https://cdn.elifesciences.org/articles/100914/elife-100914-fig5-data1-v1.zip
-
Figure 5—source data 2
Excel file containing original SDS-PAGE gels and autoradiograph photo for Figure 5b and c, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100914/elife-100914-fig5-data2-v1.zip
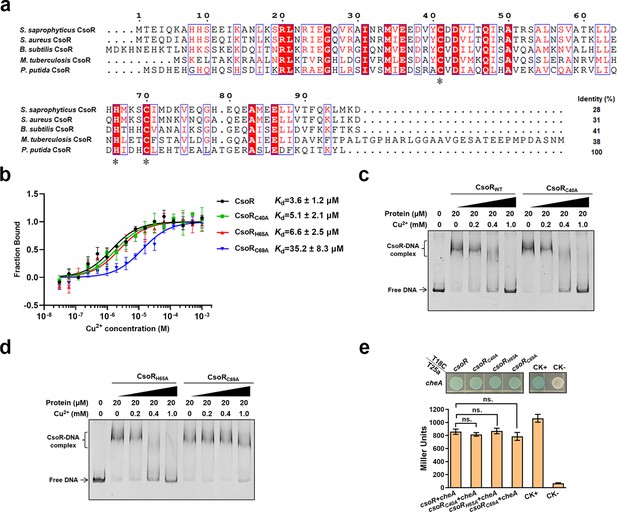
Role of the three conserved residues in the Cu2+-binding ability of CsoR.
(a) P. putida CsoR and CsoR homologs of indicated bacterial species are aligned using an online ClustalW server (https://www.genome.jp/tools-bin/clustalw). The number above the sequence represents the amino acid order of S. saprophyticus CsoR. The identical residues are shown as white on red letters. Similar residues are shown in red in blue boxes. Asterisks indicated the three conserved residues involved in Cu2+-binding. The sequence identity of CsoR homologs to the P. putida CsoR was calculated. (b) MST analysis of the interaction between wild-type and mutated CsoRs and Cu2+. Wild-type/mutated CsoR (250 nM) was incubated with increasing concentrations of Cu2+. (c and d) Effect of point mutation on the binding of CsoR to copA-I promoter in the presence of Cu2+ in the EMSA assay. Free DNA and CsoR-DNA complex are indicated. (e) Detect the interaction between CheA and point-mutated CsoR using BTH. The LacZ activities of colonies are shown below. The result is the average of three independent assays. The data represent mean values with standard deviations. ‘ns.’ represents none statistically significant between indicated strain and CK- strain analyzed by Student’s t-test.
-
Figure 5—figure supplement 1—source data 1
Excel file containing original EMSA Native-PAGE gels for Figure 5—figure supplement 1c and d.
- https://cdn.elifesciences.org/articles/100914/elife-100914-fig5-figsupp1-data1-v1.zip
-
Figure 5—figure supplement 1—source data 2
Excel file containing original EMSA Native-PAGE gels for Figure 5—figure supplement 1c and d, indicating the relevant bands and treatments.
- https://cdn.elifesciences.org/articles/100914/elife-100914-fig5-figsupp1-data2-v1.zip
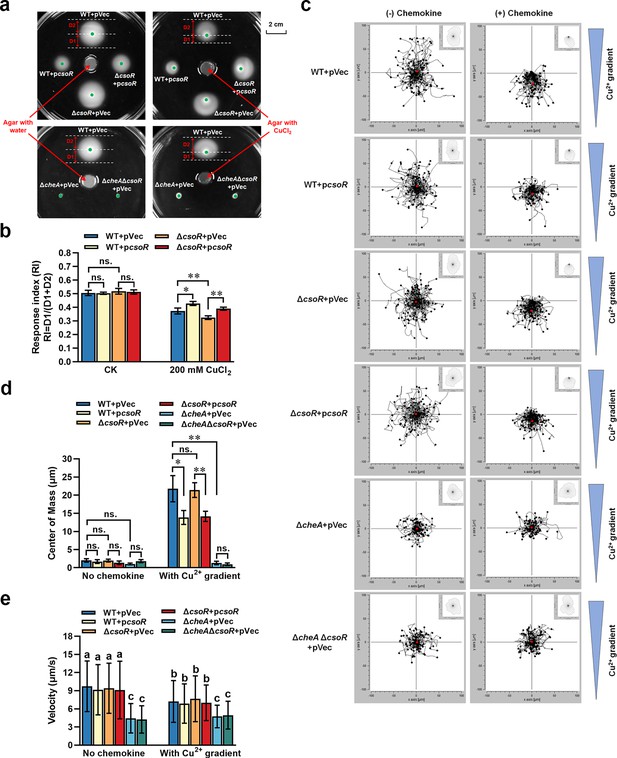
Role of CsoR in bacterial chemotaxis to copper.
(a) Chemotaxis of indicated strains in the absence and presence of copper gradient. The chemotaxis rings and chemotaxis distance (D1/D2) were indicated by arrows. The red arrows pointed at the agar plug with or without copper in the center of the plate. The green dots represented the sites where the bacteria were initially inoculated on the semisolid plate. The assay was performed with three repeats, and a representative photo was shown. (b) RI value (D1/(D1 + D2)) of indicated strains shown in panel a. (c) Aggregated trajectories of individual tested strain cells in the absence and presence of copper gradient. The tracking data presented is a composite of two experiments performed in duplicate (n=100 cells). The overall directionality of migration is depicted in the rose diagram in the upper right corner of each single-track summary. (d) Center of mass of tested strains in the presence of copper gradients. It represents the average of all single-cell endpoints. The results of panels b and d are the average of three independent assays. Error bars represent standard deviations. The asterisks represent statistically significant differences between the two indicated strains (*p<0.05, **p<0.01). ‘ns.’ represents no statistically significant between the two indicated strains. (e) Velocity analysis of indicated strains in the presence or absence of copper gradient (n=100 cells). The lowercase letters above each bar in panel e indicate significant differences (p<0.05).
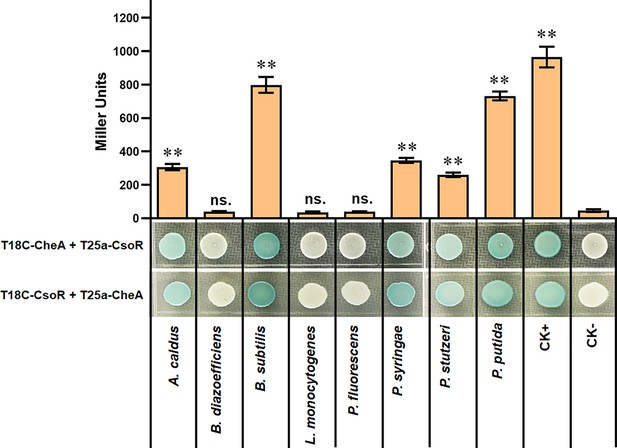
The interaction between CheA and CsoR from indicated bacterial species.
The interaction between CsoR and CheA was tested by using BTH. The LacZ activities of colonies were shown above the colonies. The results are the average of three independent assays. Error bars represent standard deviations. The asterisks above the column denote significant differences (**p<0.01) between indicated strains and CK- strain analyzed by Student’s t-test. ‘ns.’ represents none statistically significant between indicated strain and CK- strain.
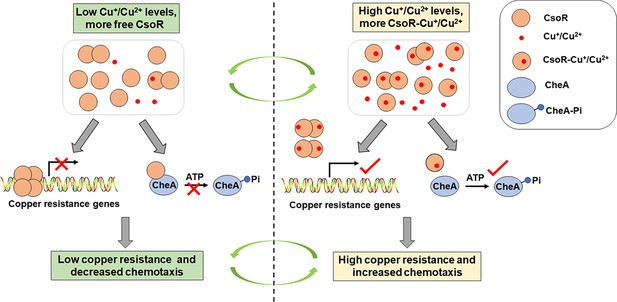
A proposed model for describing how CsoR coordinates chemotaxis and resistance to copper in P.putida.
Under low Cu+/Cu2+ levels, more none Cu+/Cu2+-binding CsoR molecules (free CsoR) exist in the cell, and the free CsoR forms tetramer and binds to promoters of copper-resistance genes (such as copA-I and copA-II), leading to repressed gene transcription and low copper resistance. Meanwhile, free CsoR interacts with CheA and inhibits its autophosphorylation activity, decreasing chemotaxis ability. In contrast, more Cu+/Cu2+-binding CsoR molecules exist under high Cu+/Cu2+ levels, and the Cu+/Cu2+-binding changes the conformation of the CsoR tetramer and releases CsoR from target promoters, leading to increased gene transcription and copper resistance. Besides, Cu+/Cu2+-binding of CsoR decreases the interaction between CsoR and CheA, which relieves the inhibition of CsoR on CheA autophosphorylation, resulting in increased chemotaxis ability.
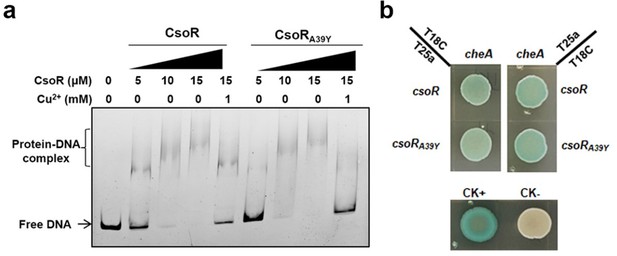
The effect of CsoR point mutation (CsoRA39Y) on the DNA-binding and Cu2+-binding abilities of CsoR.
(a) Analysis for interactions between CsoR/CsoRA39Y and copA-I promoter DNA using EMSA. The concentrations of CsoR/CsoRA39Y and Cu2+ added in each lane are shown above the gel. Free DNA and protein-DNA complexes are indicated. (b) The interaction between CsoR/CsoRA39Y and CheA was tested by BTH. Blue indicates protein-protein interaction in the colony after 60 h of incubation, while white indicates no protein-protein interaction. CK+ represents positive control, and CK- represents negative control.
Additional files
-
Supplementary file 1
Tables containing information for target proteins, strains, plasmids, and primers in this work.
(a) Target proteins identified in the pull-down assay. (b) Strains and plasmids used in this work. (c) Primers used in this work.
- https://cdn.elifesciences.org/articles/100914/elife-100914-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/100914/elife-100914-mdarchecklist1-v1.pdf