Early and delayed STAT1-dependent responses drive local trained immunity of macrophages in the spleen
Figures
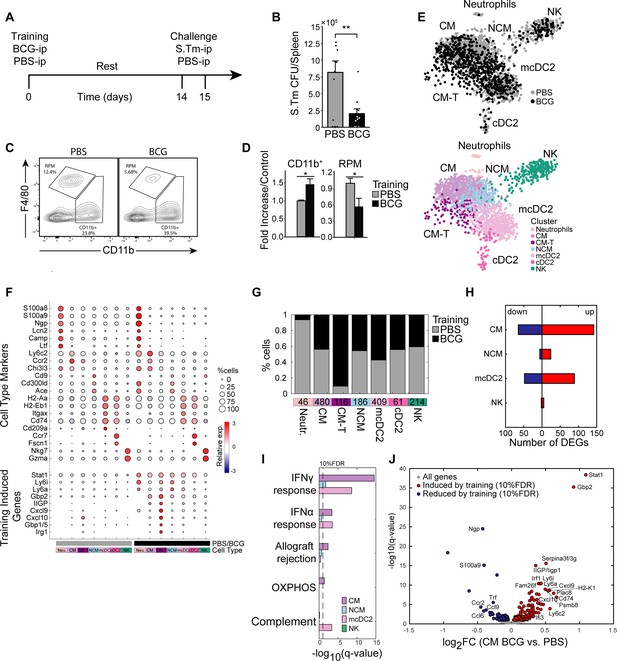
Intraperitoneal BCG results in heterologous S.Tm protection and a distinct myeloid subsets with signatures driven by STAT1.
(A) Mouse model of BCG vaccination and S.Tm challenge. (B) Splenic S.Tm CFU 24 hr post infection between control (n=11) and trained mice (n=11). (C–D) Flow cytometry plots of myeloid populations two weeks post vaccination (C) and mean percentage fold change of BCG over control for each given gated population percent from the Lin- population (D). (E) K-nearest neighbors (KNN) plot for total CD11b+ single cells sorted from control and BCG mice. Color is based on conditions, or cluster identity. (F) Cell markers and training induced genes for each subset. Size and color intensity indicates percentage of cells within a given cluster expressing the gene and average expression. (G) Proportions of monocyte subsets based on classifications in (E). (H–I) Number of DEGs in each cell subset (H), and their corresponding gene set enrichment analysis (I). (J) Volcano plot of DEGs in CM subset. Data in bar graphs are presented as mean ± SEM, with each individual point in (B) a biological repeat. Two-tailed t-test used for data in (B) and (D) (*p<0.05, **p<0.01).
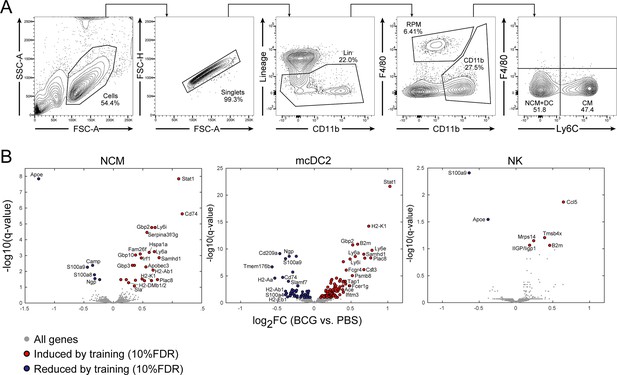
Single cell gating strategy and populations-specific DEGs.
(A) Flow cytometry gating schema for identifying MPs within the splenic myeloid subset.Gating and populations are representative of a BCG inoculated mouse two weeks post injection. Lineage (Lin) staining combines CD19 and CD3e for separation of B and T cells. (B) DEGs between PBS and BCG vaccinated mice for the sorted NCM, mcDC2, and NK populations.
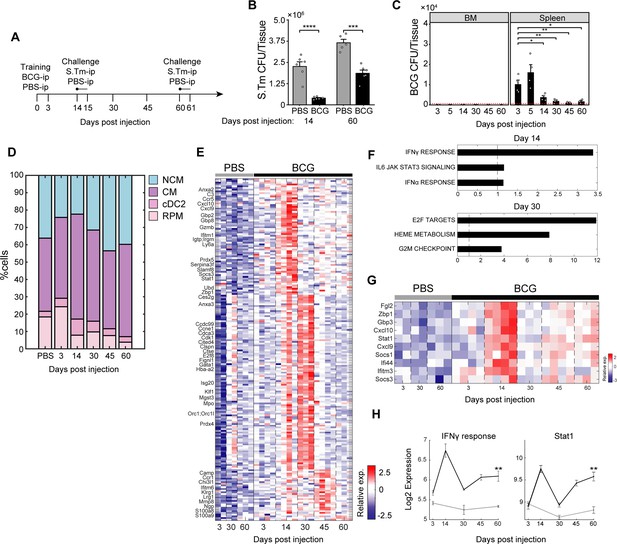
Dynamics of TI-associated subsets and signatures indicates early and delayed kinetics.
(A) Experimental setup tracking TI kinetics over a 2-month interval, including S.Tm challenge at days 14 and 60. (B) Splenic S.Tm CFU at 24 hr post infection for control and BCG mice at 14- and 60 days after vaccination (n=5–6). (C) BCG CFU from spleen and BM of BCG vaccinated mice (n=4) across time points. Red-dotted line indicates limit of detection. (D) Contribution of MP populations across time points from control (PBS) and BCG mice (days post injection). Percentage of CM, NCM, and dendritic cells calculated from flow cytometry analysis of CD11b+ population. Percentage of RPM calculated from Lin- population (control: n=3, BCG: n=4 in each time point). PBS values are the mean of all time points. (E) Heatmap of upregulated genes due to training and relative gene expression ordered according to peak expression time. (F) Gene set enrichment analysis of DEGs in days 14 and 30. (G–H) Heatmap of IFNγ response genes (G) and their average expression dynamics compared to STAT1 expression (H). Data in bar and line graphs are presented as mean ± SEM. For bar graph (B) and (C), each individual point is a biological repeat. For line graph (H), significance represents comparison between day 60 control and BCG. Heatmap rows in (E) and (G) indicate biological replicates. Two-tailed t-test used for data in (B), (C), and (H) (*p<0.05, **p<0.01, ***p<0.005, ****p<0.001).
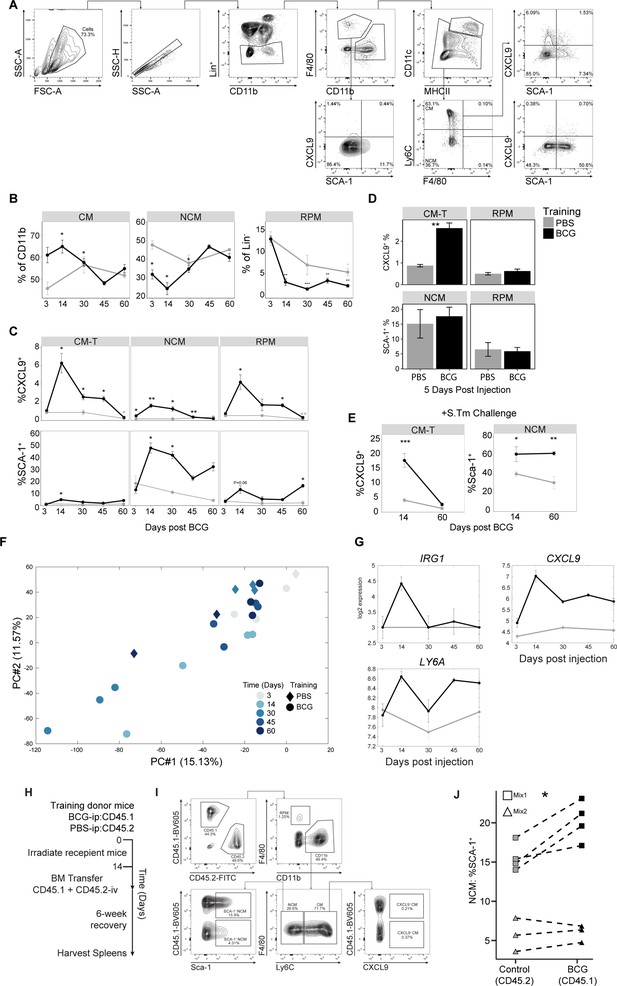
Kinetics gating strategy and marker and gene expression across time and populations.
(A) Flow cytometry gating schema for identifying MPs and TI-associated markers (including CXCL9+ CMs [CM-Ts] and Sca-1) within the splenic myeloid subset.Gating and populations are representative of a BCG inoculated mouse 2 weeks post injection. Lineage staining combines CD19, CD3e, Ly6g, and NK1.1 for separation of B, T, neutrophil, and NK cells respectively. (B–C) Percentage of MP populations (B) and frequency of CM-Ts and SCA-1 + cells (C) from PBS (gray) and BCG (black) mice. CM, NCM, and dendritic cell ratio calculated from CD11b+population. RPM ratio calculated from Lin- population (control: n=2–3, BCG: n=4). (D) Frequency of CM-Ts and SCA-1 + cells 5 days post PBS (gray) or BCG (black) vaccination (control: 3, BCG = 5). (E) Frequency of CM-Ts and SCA-1 +NCMs 24 hours post S.Tm challenge at 2 or 8 weeks after PBS (gray) or BCG (black) vaccination (PBS: n=6, BCG: n=6). (F) PCA projection of all bulk splenic conditions onto the space of the two leading principal components based on all expressed genes due to training. Conditions include each time point (control: n=2–3, BCG: n=3–4). Shade intensity represents time point, diamond (PBS) or circle (BCG). (G) Log2 expression of selected training-associated genes over time (control: n=2–3, BCG: n=3–4). (H) Experimental setup for tracking expression of training markers inherited from trained BM progenitors. After initial inoculation of PBS-ip (n=2) or BCG-ip (n=2), mice were sacrificed, and BM extracted and mixed for each pair of donors. (I) Flow cytometry gating schema for identifying MPs and TI-associated markers (CXCL9 and Sca-1) across CD45.1/CD45.2 donors. (J) NCM fraction expressing Sca-1 from cells derived from control (CD45.2) or BCG (CD45.2) mice (n=7). Square or triangle represent mice who received combined BM from donor set one or two. Significance between control (CD45.2) and BCG (CD45.1) is indicated. Data in (B–E) are presented as mean ± SEM. For line graphs (B) and (C), significance represents each time point against PBS at day 3. Two-tailed t-test used for data in (B–E) paired t-test for J (*p≥0.05, **p≥0.01, ***p≥0.005, ****p≥0.001).
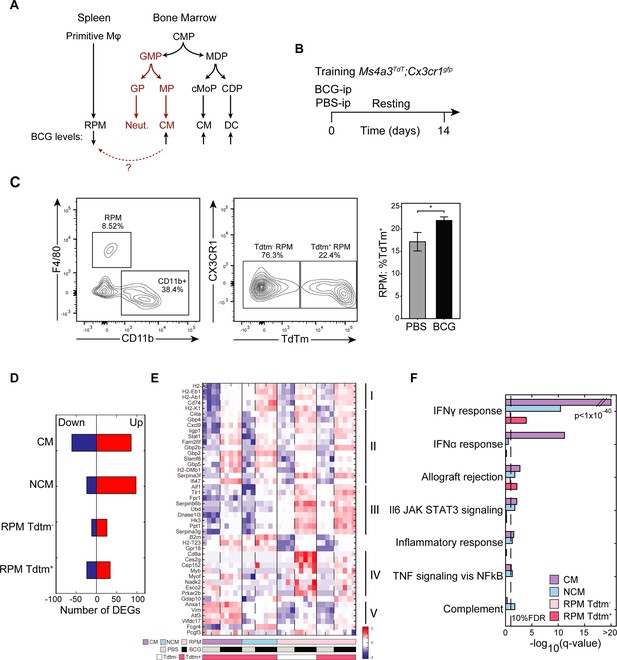
RPM niche is replenished by recruited trained monocytes and by local training of tissue-resident populations.
(A) Scheme representing known myeloid differentiation pathways and potential trans-differentiation of trained CM to RPM. (B) Mouse model to track contribution of TI-associated signatures in local and recruited MP populations with lineage tracing. (C–D) Flow cytometry analysis of TdTm+ or Tdtm- Ly6C+ MPs and RPM and quantification of RPM TdTm+ subset (n=3). (D–F) Number of DEGs of each sorted population (D), heatmap of normalized log2 expression from TI-associated DEGs specific to trained RPM populations and gene set enrichment analysis of DEGs in each sorted population (F). Data in bar graphs are presented as mean ± SEM. Heatmap rows in (E) indicate biological replicates. Two-tailed t-test used for data in (C) (*p<0.05).
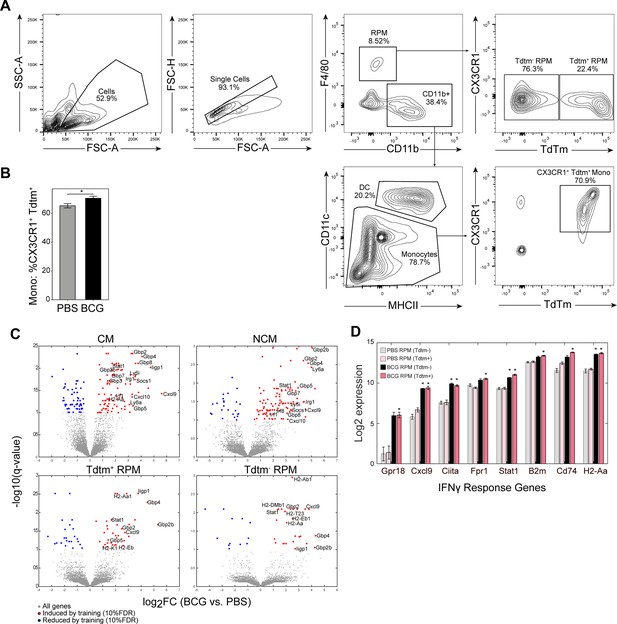
Lineage tracing gating strategy and origin-specific DEGs.
(A) Flow cytometry gating schema for identification of local and recruited MPs in the spleen.Gating and populations are representative of a BCG inoculated mouse 2 weeks post injection. (B) Percent of splenic monocytes double positive for CX3CR1 and TdTomato between PBS (gray) and BCG (black) vaccinated mice (PBS: n=4, BCG: n=5). (C) DEGs between PBS and BCG vaccinated mice for the bulk sorted CM, NCM, and Tdtm +/-RPM. (D) Expression of genes enriched within the IFNy response term across Tdtm +/-RPM. Data in (B) are presented as mean ± SEM.
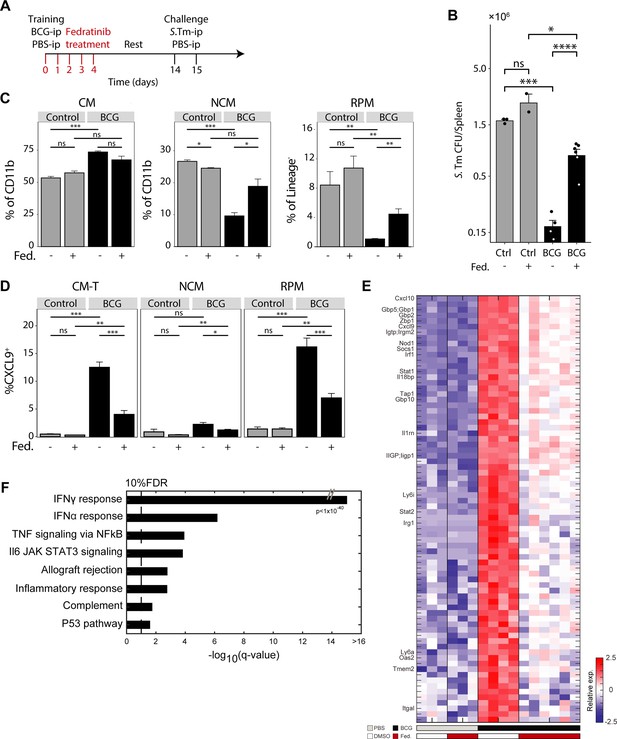
Transient IFNγ-STAT1 inhibition prevents TI signatures and splenic infection resistance.
(A) Mouse model of BCG vaccination with early interferon inhibition using the Fedratinib inhibitor. (B) Splenic S.Tm CFU for control and BCG mice, with and without Fedratinib inhibitor, 24 hr post infection (n=2–6).(C) MP populations from control (gray) and BCG (black) mice, with or without Fedratinib inhibition. Percentage of CM and NCM cells calculated from CD11b+ population. Percentage of RPM calculated from Lin- population. (D) Percentage of CXCL9+ CM-T, NCM, and RPM populations from control (gray) and BCG (black) mice, with or without Fedratinib inhibition (control: n=3, BCG: n=4, control +Fedratinib: n=3, BCG +Fedratinib: n=6). (E) Heatmap of normalized log2 expression of DEGs across naive and training conditions. (F) Gene set enrichment analysis of DEGs from E. Data in bar graphs are presented as mean ± SEM. For bar graph (B) each individual point is a biological repeat. Heatmap rows in E indicate biological replicates. Two-tailed t-test used for data in (B), (C), and (D) (*p<0.05, **p<0.01, ***p<0.005, ****p<0.001).
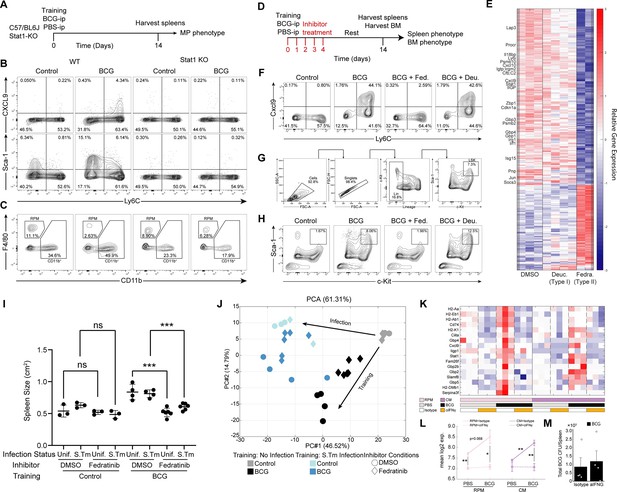
Training inhibition with Stat-1 KO mice and across interferon inhibition strategies.
(A) Mouse model to detect training markers in BCG-vaccinated WT and STAT1-KO mice. (B) CXCL9 and Sca-1 expression in MPs between control and BCG mice in WT or STAT1-KO background. (C) CD11b+ and RPM populations between control and BCG mice in WT or STAT1-KO background. (D) Mouse model of BCG vaccination with early interferon inhibition using the small molecule inhibitor Fedratinib targeting Type-II IFN, or Deucravacitinib targeting Type-I IFN. (E) Heatmap of log2 fold change for DEGs across inhibitor conditions. (F) CXCL9 expression in MPs across inhibitor conditions. (G) Flow cytometry gating strategy used for identification of BM LSK HSCs. Gating is representative of a BCG inoculated mouse 2 weeks post injection. Lineage staining combines CD4, CD8, B220, Ter119, Gr1, B220, and CD11b for separation of T, B, erythroid, granulocyte, and myeloid cells respectively. (H) Flow cytometry plot of LSK expansion within the BM across inhibitor conditions. (I) Two-dimensional spleen area from across experimental conditions including training, Fedratinib inhibitor, and S.Tm infection (n=3–6). Significance between inhibitor and infection conditions is indicated. (J) PCA projection of all samples onto the space of the two leading principal components based on all DEGs across training, inhibitor, and infection conditions (two-sample t-test, 5% FDR). The percentage of variance explained by each PC is indicated at the PC axes. Color is indicative of control or BCG mice before and after S.Tm challenge. Circle or diamond shape represent treatment with DMSO or Fedratinib. (K–L) Heatmap of STAT-1 signature genes (K) and their mean log2 expression across training conditions and antibody treatment (L). (M) Total BCG CFU from spleens from isotype and α-IFNγ treated mice 2 weeks post vaccination (n=4). Data in (I) and (M) are presented as mean ± SEM. Heatmap rows in (E) and (K) indicate biological replicates. Two-tailed t-test used for data in (I) and (L) (*p≥0.05, **p≥0.01, ***p≥0.005, ****p≥0.001).
Additional files
-
Supplementary file 1
Supplementary tables indicating significant DEGs.
(a) Training induced DEGs from single-cell experiment. For each population (CM, NCM, mcDC, and NK cells), significantly differentially expressed genes (10% FDR) between naive and trained cells are presented, with log-ratio values, and corresponding p-value and q-value.(b) Training induced DEGs from the kinetics experiment. Total splenocyte log2-ratio expression from both naïve (PBS) and trained (BCG) vaccinated mice at days 3, 14, 30, 45, and 40 days after vaccination. Replicates are marked at the end of the sample name.(c) Significantly expressed DEGs (10% FDR) between naïve and trained splenic CM, NCM, and Tdtm+/- RPM. Log-ratio values, and corresponding p-value and q-values presented.(d) Training induced significant DEGs (10% FDR) in total splenocytes, between naïve and trained mice, with and without Type I and II interferon inhibition. Values are log2-ratio expression per replicate. Replicates are marked at the end of the sample name.(e) Training induced significant DEGs (10% FDR) in total splenocytes, between naive and trained mice, with and without Fedratinib inhibition. Values are log2-ratio expression per replicate. Replicates are marked at the end of the sample name.
- https://cdn.elifesciences.org/articles/100922/elife-100922-supp1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/100922/elife-100922-mdarchecklist1-v1.docx





