A bacterial regulatory uORF senses multiple classes of ribosome-targeting antibiotics
Figures
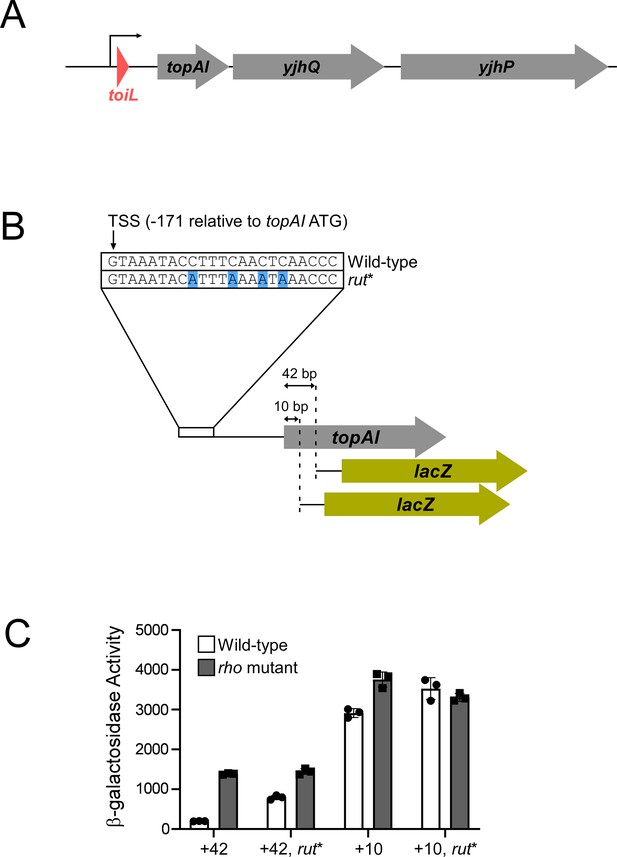
Rho-dependent transcription termination within the topAI gene.
(A) Schematic representation of the E. coli topAI-yjhQ-yjhP operon, including the toiL uORF. (B) Schematic representation of topAI-lacZ transcriptional reporter fusions, indicating the position of the transcription start site (TSS) with respect to the topAI start codon, and the mutation in the putative rut. (C) β-galactosidase activity of topAI-lacZ transcriptional fusions in wild-type (strain AMD054) or rho mutant (R66S amino acid substitution in Rho; strain GB004) cells. Reporter fusions with wild-type sequence or mutation of the predicted rut (rut*) extend to position + 42 or +10 of topAI (wild-type + 42, plasmid pGB217; wild-type + 10, plasmid pGB215; rut* + 42, plasmid pGB306; rut* +10, plasmid pGB305). Error bars represent ±1 standard deviation from the mean (n=3).

topAI expression is induced in response to translation stress.
(A) Schematic representation of topAI-lux translational reporter fusions, indicating the position of the toiL start codon with respect to the topAI start codon. (B) Luciferase activity of the wild-type topAI-lux translational reporter fusion (pGB202) in ΔtopAI-yjhQ cells (strain GB001) expressing either wild-type 23S rRNA (plasmid pGB322) or ΔT1917 23S rRNA (plasmid pGB318) in trans. Activity was measured 4 hours post-induction with propionate. (C) Luciferase activity of wild-type and mutant topAI-lux translational reporter fusions in ΔtopAI-yjhQ cells (strain GB001) grown in the presence of ribosome-targeting antibiotics. Cells were grown in LB medium to an OD600 of ~1.0. Indicated samples were treated with tetracycline (0.5 μg/ml), spectinomycin (90 μg/ml), retapamulin (4 μg/ml), erythromycin (100 μg/ml), tylosin (400 μg/ml), or chloramphenicol (1 μg/ml). Luminescence was measured 90 min after antibiotic treatment. Error bars represent ±1 standard deviation from the mean (n=3).
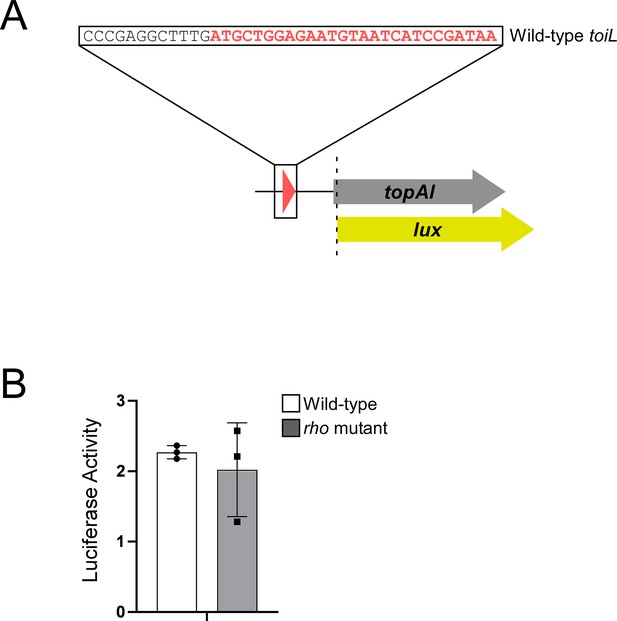
topAI is translationally repressed.
(A) Schematic representation of the topAI-lux translational reporter fusion. (B) Luciferase activity of the topAI-lux translational fusion (plasmid pGB202) in ΔtopAI-yjhQ cells (‘wild-type’; strain GB001) or in ΔtopAI-yjhQ rho mutant (R66S amino acid substitution in Rho; strain GB004). Error bars represent ±1 standard deviation from the mean (n=3).
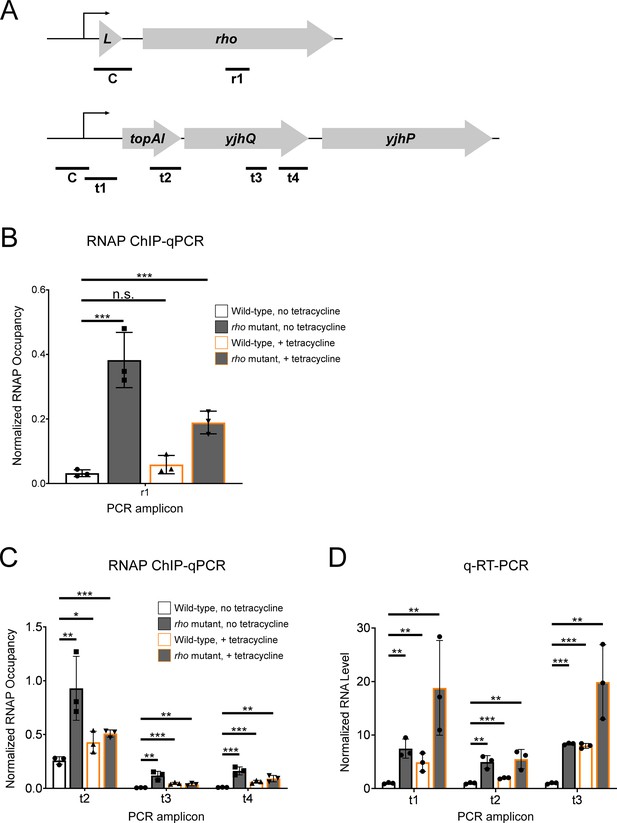
Tetracycline treatment reduces premature Rho-dependent transcription termination within topAI-yjhQ and increases abundance of the associated mRNA.
(A) Schematic of the rhoL-rho and topAI-yjhQP operons showing the positions of PCR amplicons used for ChIP-qPCR and/or qRT-PCR. (B) Occupancy of RNAP measured by ChIP-qPCR in the rho gene. Occupancy values are normalized to those in rhoL (control PCR amplicon ‘C’ in panel A). Data are shown for wild-type (strain MG1655) or rho mutant (R66S amino acid substitution in Rho; strain CRB016) cells grown in the presence or absence of tetracycline, as indicated. The x-axis label corresponds to the PCR amplicon shown in panel A. (C) Occupancy of RNAP measured by ChIP-qPCR at positions across the topAI-yjhQ transcript. Occupancy values are normalized to those in the topAI upstream region (control PCR amplicon ‘C’ in panel A). (D) RNA levels measured by qRT-PCR at positions across the topAI-yjhQ transcript, normalized to the mreB gene. Error bars represent ±1 standard deviation from the mean (n=3). Statistical significance is indicated as follows: n.s., not significant (p>0.05), *p<0.05, **p<0.01, ***p<0.001.
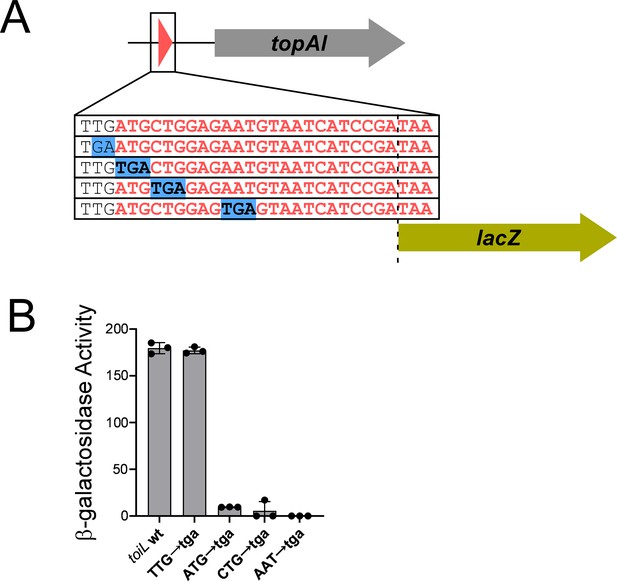
A uORF, toiL, is located within the topAI 5’ upstream region.
(A) Schematic representation of toiL-lacZ translational reporter fusions, indicating mutations upstream and within the toiL uORF. (B) β-galactosidase activity of the wild-type toiL-lacZ translational reporter fusion (wt; plasmid pGB164), and fusions with mutations immediately upstream of toiL (TTG→tga; plasmid pGB201), at the toiL start codon (ATG→tga; plasmid pGB200), second codon (CTG→tga; plasmid pGB197), or fourth codon (AAT→tga; plasmid pGB196), in cells with wild-type topAI-yjhQP (strain AMD054). Error bars represent ±1 standard deviation from the mean (n=3).

ToiL conservation across Enterobacteriaceae species.
Predicted ToiL sequences were aligned with ClustalO (Madeira et al., 2022). Asterisks indicate fully conserved positions.
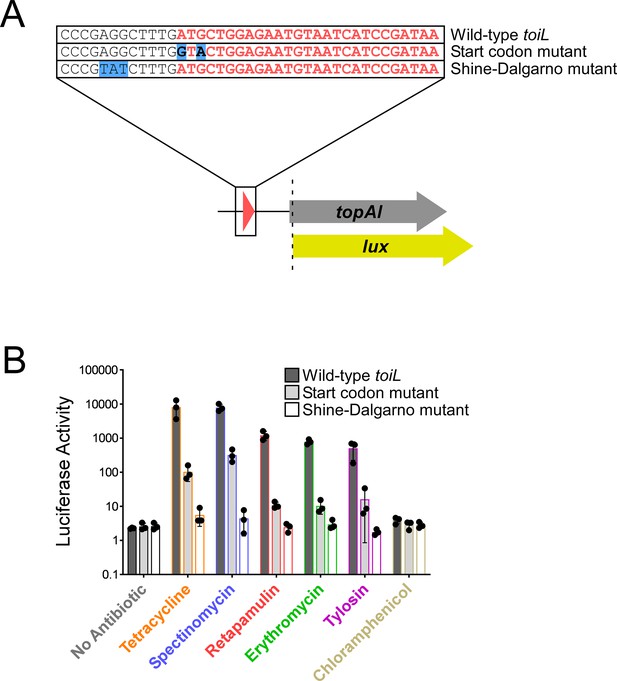
toiL translation is required for induction of topAI expression by ribosome-targeting antibiotics.
(A) Schematic representation of topAI-lux translational reporter fusions, indicating mutations in the start codon and Shine–Dalgarno sequence of the toiL uORF. (B) Luciferase activity of wild-type and mutant topAI-lux translational reporter fusions in ΔtopAI-yjhQ cells (strain GB001) grown in the presence of ribosome-targeting antibiotics. Cells were grown as indicated in Figure 2C. Dark gray bars show data for the wild-type topAI-lux translational reporter fusion (plasmid pGB202); light gray bars show data for the topAI-lux translational reporter fusion with a mutated toiL start codon (plasmid pGB313); white bars show data for the topAI-lux translational reporter fusion with a mutated toiL Shine–Dalgarno sequence (pGB366). Note that data for the wild-type topAI-lux translational reporter fusion are identical to those in Figure 2C and are included here for reference. Error bars represent ±1 standard deviation from the mean (n=3).
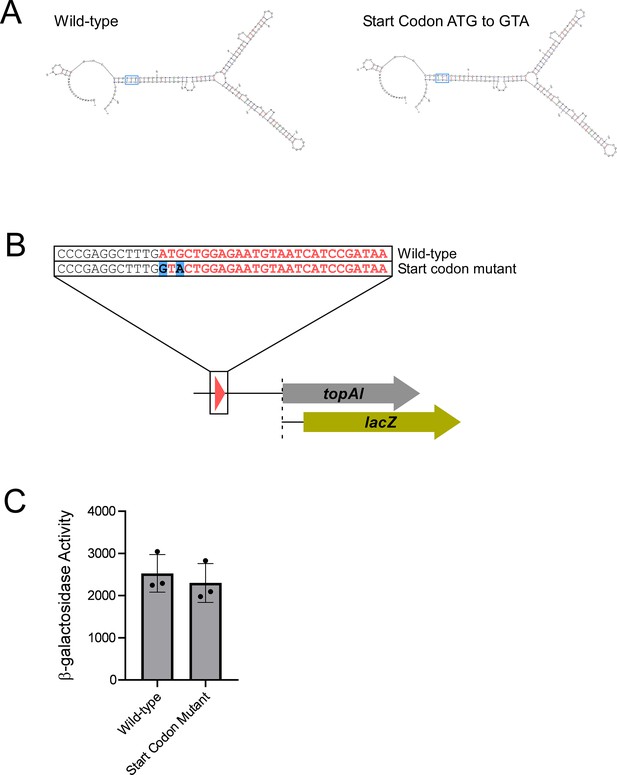
Mutation of the toiL start codon does not alter predicted RNA secondary structure and does not lead to Rho-dependent transcription termination in the topAI 5’ upstream region.
(A) Predicted minimum free-energy secondary structures of the topAI 5’ UR for wild-type sequence and sequence where the toiL start codon (ATG) is mutated (GTA). (B) Schematic representation of topAI-lacZ transcriptional reporter fusions indicating the start codon mutation of toiL. (C) β-galactosidase activity of the wild-type topAI-lacZ transcriptional reporter (plasmid pGB182) and the toiL start codon mutant reporter (plasmid pGB297) in ΔtopAI-yjhQ cells (strain GB001). Error bars represent ±1 standard deviation from the mean (n=3).
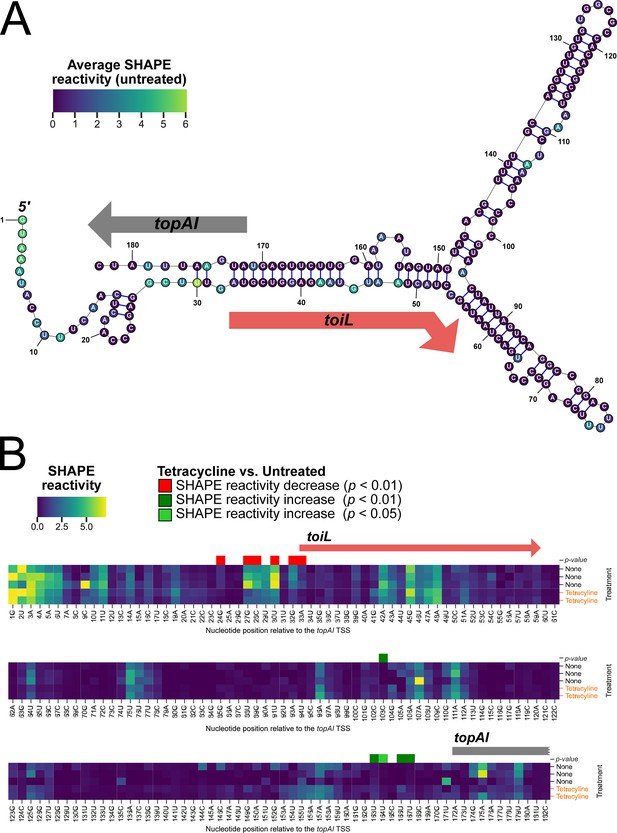
Structural changes in the topAI 5′ upstream region induced by tetracycline treatment.
(A) Predicted RNA secondary structure of the topAI 5’ UR, indicating in-cell SHAPE reactivities of each base from an untreated sample. Reactivity scores are indicated on a color gradient. The position of the toiL uORF is indicated by a red arrow, and the position of the start of topAI is indicated by a gray arrow. Numbers indicate position relative to the mRNA 5’ end. (B) In-cell SHAPE reactivities of each base from three replicates of untreated samples and two replicates of tetracycline-treated samples. SHAPE reactivity values are indicated on the same color gradient as panel A. Nucleotide positions with statistically significant differences (p<0.01 or 0.05, as indicated) in SHAPE reactivity between untreated and tetracycline-treated samples are indicated by red (lower reactivity in tetracycline-treated samples) or green (higher reactivity in tetracycline-treated samples) squares.
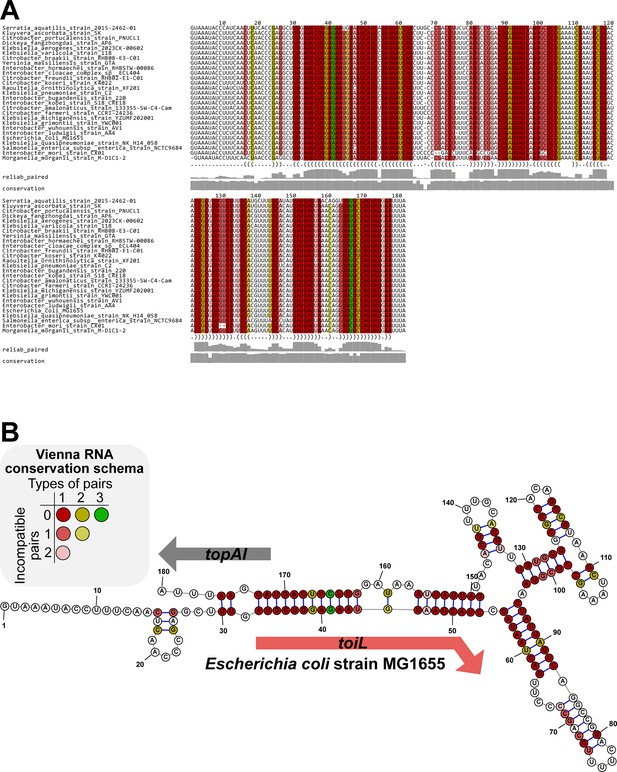
Predicted RNA secondary structure for the topAI upstream region based on sequence conservation.
(A) PETfold alignment of topAI 5’ UR sequences from selected species. Predicted base pairing interactions are represented in a dot-bracket format below the alignment. Reliability of base pairing (reliab_paired) and sequence conservation scores are represented as grey bars (0–1 scale). (B) Representation of the PETfold-predicted topAI 5’ UR RNA consensus structure. Nucleotide colors in both panels follow the Vienna RNA conservation coloring schema (Bernhart et al., 2008) and represent the number of compatible or incompatible nucleotide pairs for each position, as indicated.
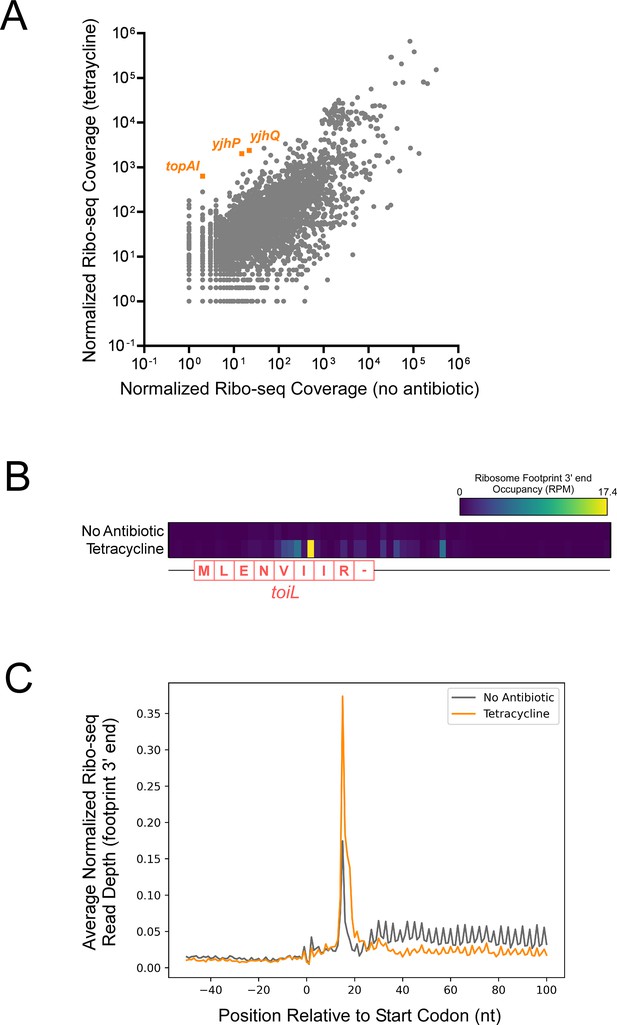
Tetracycline stalls ribosomes on start codons.
(A) Normalized Ribo-seq coverage for all annotated ORFs for cells (strain MG1655) grown ± tetracycline. (B) Heatmap showing normalized Ribo-seq coverage in the region around the toiL uORF in untreated cells (no antibiotic) or tetracycline-treated cells. The color indicates the sequence-read coverage (reads per million; RPM) for RNA fragment 3’ ends that are presumed to represent the downstream edge of ribosome footprints. The toiL uORF position and encoded amino acid sequence is indicated. (C) Average of normalized Ribo-seq coverage for the regions around start codons for all annotated ORFs, for untreated cells (gray line; no antibiotic) or tetracycline-treated cells (orange line). Sequence-read coverage was calculated for RNA fragment 3’ ends that are presumed to represent the downstream edge of ribosome footprints.
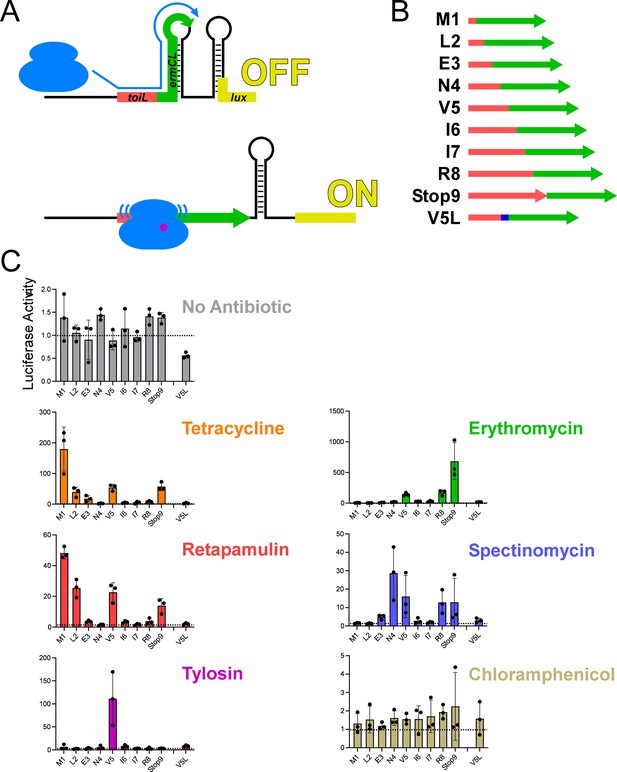
Ribosome stalling at the topAI leader in vivo.
(A) Schematic showing the toiL-ermCL-lux stalling reporter in the translationally inactive conformation (‘OFF’) that is expected in the absence of ribosome stalling, and the translationally active conformation (‘ON’) that is expected when ribosomes stall within toiL sequence close to the junction of toiL with ermCL. (B) Schematic showing toiL-ermCL stalling reporters where toiL is progressively extended by one codon at the 3’ end and fused to part of ermCL. toiL sequence is indicated by red rectangles/arrow; ermCL sequence is indicated by green arrows. The last indicated stalling reporter extends to position 5 of toiL but has a Val5→Leu substitution (blue). (C) Luciferase activity of the toiL-ermCL-lux stalling reporters (plasmids pGB323, pGB346, pGB325, pGB326, pGB324, pGB328, pGB329, pGB307, pGB347, and pGB327) in ΔtopAI-yjhQ cells (strain GB001). Cells were grown to an OD600 of ~1.0 and treated with the indicated ribosome-targeting antibiotics at the same concentrations as in Figure 2C. Luminescence was measured three hours post-treatment. Horizontal dashed lines indicate luminescence activity of 1, to facilitate comparison between panels with different y-axis scales. Error bars represent ±1 standard deviation from the mean (n=3).
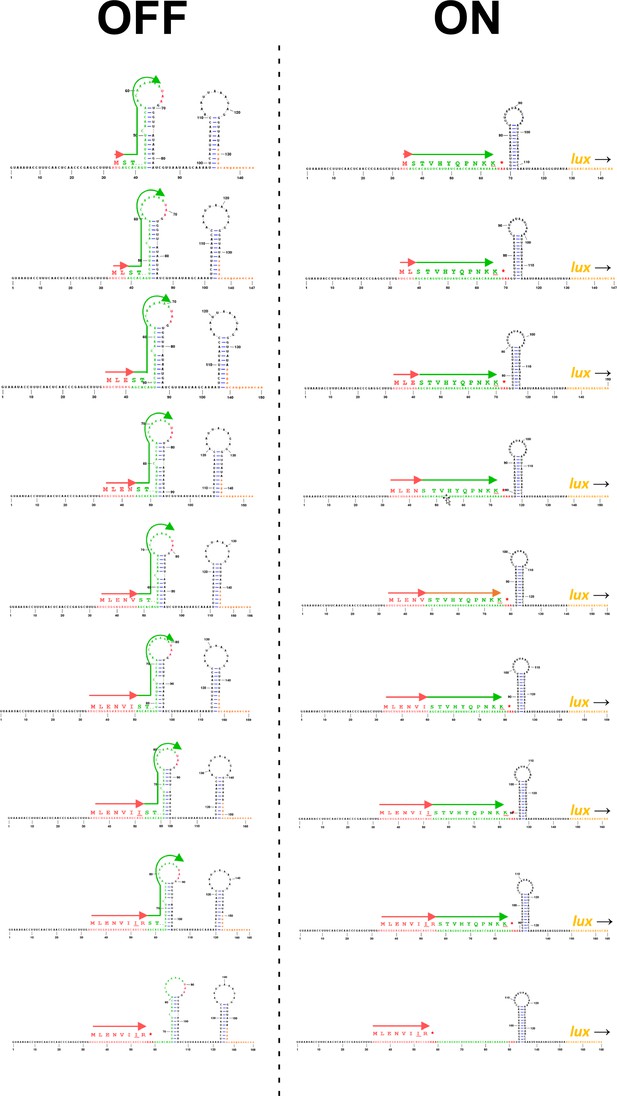
Predicted RNA structures for translationally inactive and active conformations of toiL stalling reporters.
Predicted RNA structures are shown for toiL-ermCL-lux stalling reporters where increasing lengths of toiL are fused to part of ermCL. toiL is indicated by red arrows. ermCL sequence is indicated by green arrows. Pausing of ribosomes within toiL, close to the junction with ermCL sequence, is expected to promote formation of the active (‘ON’) conformation, whereas the absence of ribosome stalling within toiL is expected to promote formation of the inactive (‘OFF’) conformation.
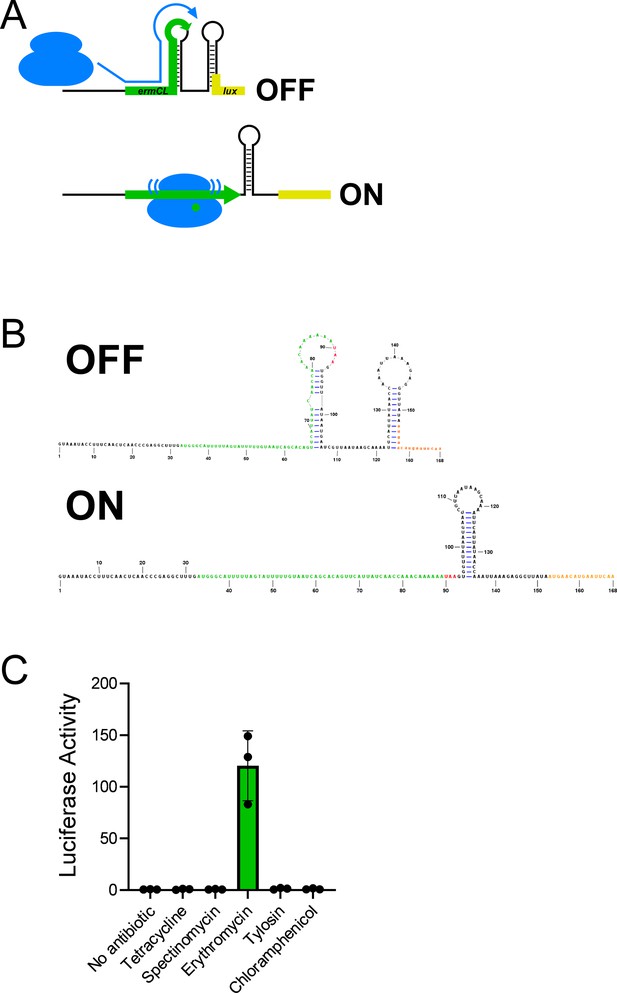
Expression of an ermCL stalling reporter is specifically induced by erythromycin.
(A) Schematic showing the ermCL-lux stalling reporter in the translationally inactive conformation (‘OFF’) that is expected in the absence of erythromycin, when ribosomes do not stall within the ermCL uORF; and the translationally active conformation (‘ON’) that is expected when ribosomes stall within ermCL in the presence of erythromycin. (B) Predicted RNA secondary structures of the inactive (‘OFF’) and active (‘ON’) conformations of the ermCL-lux stalling reporter. (C) Luciferase activity of the ermCL-lux stalling reporter (plasmid pGB308) in ΔtopAI-yjhQ cells (strain GB001) treated with the indicated ribosome-targeting antibiotics. Cells were grown to an OD600 of ~1.0 and treated with the indicated ribosome-targeting antibiotics at the same concentrations as in Figure 2C. Luminescence was measured 3 hours post-treatment. Error bars represent ±1 standard deviation from the mean (n=3).

Model for regulation of topAI expression.
Schematic showing a model for regulation of topAI expression. In the repressed state, a hairpin forms between the toiL sequence and the ribosome-binding site of topAI, repressing translation, which in turn promotes premature Rho-dependent transcription termination within the topAI gene. Ribosomes do not stall within toiL, so there is minimal perturbation of the repressive hairpin. By contrast, when cells are treated with certain ribosome-targeting antibiotics such as tetracycline, ribosomes stall within toiL in a sequence-specific manner, preventing formation of the repressive hairpin.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | MG1655 (WT) | This paper | N/A | See Supplementary file 1E for derivatives |
| Antibody | Mouse monoclonal lnti-E. coli RNA Polymerase β | BioLegend | 663003, clone: NT63; RRID:AB_2564516 | ChIP (1:800) |
| Recombinant DNA reagent | pAMD-BA-lacZ | Stringer et al., 2014 | N/A | See Supplementary file 1E for derivatives |
| Recombinant DNA reagent | pPro24 | Addgene | RRID:Addgene_17805 | See Supplementary file 1E for derivatives |
| Recombinant DNA reagent | pET-PesaR-lux | Addgene | RRID:Addgene_47802 | See Supplementary file 1E for derivatives |
| Recombinant DNA reagent | pCS-PesaR-lux | Addgene | RRID:Addgene_47655 | See Supplementary file 1E for derivatives |
| Sequence-based reagent | Primers and geneblocks | IDT | N/A | See Supplementary file 1E for derivatives |
| Chemical compound, drug | Tetracycline | Sigma-Aldrich | T3383 | |
| Chemical compound, drug | Spectinomycin | Sigma-Aldrich | S4014 | |
| Chemical compound, drug | Erythromycin | Sigma-Aldrich | E5389 | |
| Chemical compound, drug | Retapamulin | Sigma-Aldrich | CDS023386 | |
| Chemical compound, drug | Tylosin | Sigma-Aldrich | T6134 | |
| Chemical compound, drug | Chloramphenicol | Sigma-Aldrich | C0378 | |
| Chemical compound, drug | Gentamicin | Sigma-Aldrich | G1272 | |
| Chemical compound, drug | Hygromycin B | Millipore | 400052 | |
| Chemical compound, drug | Sodium propionate | Sigma-Aldrich | P1880 | |
| Chemical compound, drug | 1M7 | MedChem Express | HY-D0913 | |
| Commercial assay or kit | Quick-RNA Miniprep Kit | Zymo Research | R1054 | |
| Commercial assay or kit | MultiScribe Reverse Transcriptase | Invitrogen | 4311235 | |
| Commercial assay or kit | SuperScript III Reverse Transcriptase | Invitrogen | 18080093 | |
| Commercial assay or kit | DNeasy Blood & Tissue Kit | QIAGEN | 69504 | |
| Commercial assay or kit | NEBuilder HiFi DNA Assembly Master Mix | NEB | E2621S | |
| Other | SphI-HF | NEB | R3182S | Restriction enzyme used for cloning plasmids |
| Other | BamHI-HF | NEB | R3136S | Restriction enzyme used for cloning plasmids |
| Other | HindIII-HF | NEB | R3104S | Restriction enzyme used for cloning plasmids |
| Other | XhoI-HF | NEB | R0146S | Restriction enzyme used for cloning plasmids |
| Other | XbaI-HF | NEB | R0145S | Restriction enzyme used for cloning plasmids |
| Other | EcoRI-HF | NEB | R3101S | Restriction enzyme used for cloning plasmids |
| Other | NheI-HF | NEB | R3131S | Restriction enzyme used for cloning plasmids |
| Other | SHAPE-seq reagents | Watters et al., 2016a | N/A | Used for experiments in Figure 6 |
| Other | Ribo-seq reagents | Stringer et al., 2021 | N/A | Used for experiment in Figure 7 |
| Software, algorithm | Python | Python Language Reference | RRID:SCR_008394 | v3.7; https://www.python.org/ |
| Software, algorithm | seaborn | https://doi.org/10.21105/joss.03021 | RRID:SCR_018132 | v0.11.1; https://seaborn.pydata.org/index.html |
| Software, algorithm | R | The R Project | RRID:SCR_001905 | v4.1.2; https://www.r-project.org/ |
| Software, algorithm | DESeq2 | Love et al., 2014 | RRID:SCR_015687 | v1.34.0; https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Software, algorithm | CLC Genomics Workbench | QIAGEN | v8.5.1; SNP calling | |
| Software, algorithm | spats | Watters et al., 2016a; Trapnell et al., 2017 | v1.0.2; https://luckslab.github.io/spats/ | |
| Software, algorithm | cutadapt | https://doi.org/10.14806/ej.17.1.200 | RRID:SCR_011841 | v0.12.8; https://cutadapt.readthedocs.io/en/stable/ |
| Software, algorithm | bowtie | https://doi.org/10.1186/gb-2009-10-3-r25 | RRID:SCR_005476 | v0.12.8; http://bowtie.cbcb.umd.edu/ |
| Software, algorithm | Rockhopper | McClure et al., 2013 | v2.0.3; https://cs.wellesley.edu/~btjaden/Rockhopper/index.html | |
| Software, algorithm | ClustalO | Madeira et al., 2022 | RRID:SCR_001591 | https://www.ebi.ac.uk/jdispatcher/msa/clustalo |
| Software, algorithm | Prism | GraphPad | RRID:SCR_002798 | v9.4.1 |
| Software, algorithm | PETfold | Seemann et al., 2011 | v2.2; https://rth.dk/resources/petfold/submit.php | |
| Software, algorithm | Mfold | Zuker, 2003 | RRID:SCR_008543 | v4.7; https://www.unafold.org/mfold/applications/rna-folding-form.php |
| Software, algorithm | RNAStructure | https://doi.org/10.1186/1471-2105-11-129 | RRID:SCR_017216 | v6.1; https://rna.urmc.rochester.edu/RNAstructure.html |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/101217/elife-101217-mdarchecklist1-v1.pdf
-
Supplementary file 1
Supplementary tables.
- https://cdn.elifesciences.org/articles/101217/elife-101217-supp1-v1.xlsx





